7YEN
 
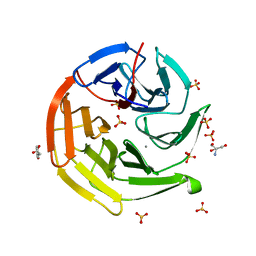 | |
7WN0
 
 | | Structure of PfENT1(Y190A) in complex with nanobody 19 | | Descriptor: | Equilibrative nucleoside/nucleobase transporter, nanobody19 | | Authors: | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | Deposit date: | 2022-01-17 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
7YDQ
 
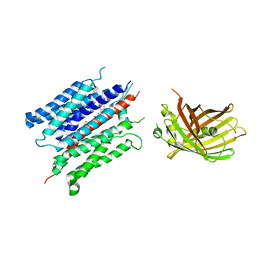 | | Structure of PfNT1(Y190A)-GFP in complex with GSK4 | | Descriptor: | 5-methyl-N-[2-(2-oxidanylideneazepan-1-yl)ethyl]-2-phenyl-1,3-oxazole-4-carboxamide, Nucleoside transporter 1,Green fluorescent protein | | Authors: | Wang, C, Yu, L.Y, Li, J.L, Ren, R.B, Deng, D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
4EOT
 
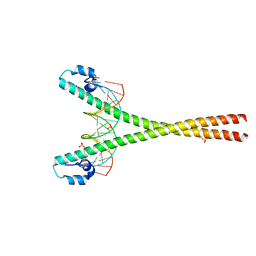 | | Crystal structure of the MafA homodimer bound to the consensus MARE | | Descriptor: | 5'-D(*CP*CP*CP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*GP*CP*AP*CP*CP*G)-3', 5'-D(*CP*CP*GP*GP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*GP*G)-3', SULFATE ION, ... | | Authors: | Lu, X, Guanga, G, Wan, C, Rose, R.B. | | Deposit date: | 2012-04-15 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.855 Å) | | Cite: | A Novel DNA Binding Mechanism for maf Basic Region-Leucine Zipper Factors Inferred from a MafA-DNA Complex Structure and Binding Specificities.
Biochemistry, 51, 2012
|
|
7V6G
 
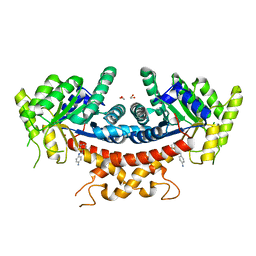 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase mutation C157S with CN39 | | Descriptor: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION, ... | | Authors: | Cao, H, Huang, Y, Chen, H, Wan, C, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
6XGF
 
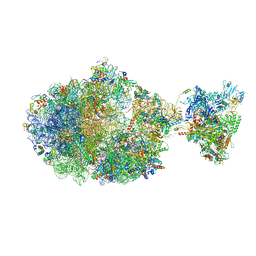 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 30 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-17 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X9Q
 
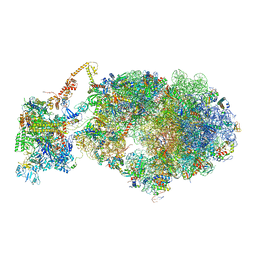 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 27 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XII
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 24 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-20 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X6T
 
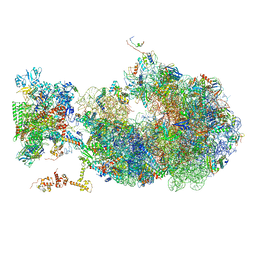 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B1 (TTC-B1) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XIJ
 
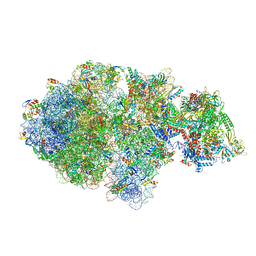 | | Escherichia coli transcription-translation complex A (TTC-A) containing an 24 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-20 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6OHW
 
 | | Structural basis for human coronavirus attachment to sialic acid receptors. Apo-HCoV-OC43 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike surface glycoprotein, ... | | Authors: | Tortorici, M.A, Walls, A.C, Lang, Y, Wang, C, Li, Z, Koerhuis, D, Boons, G.J, Bosch, B.J, Rey, F.A, de Groot, R, Veesler, D. | | Deposit date: | 2019-04-07 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human coronavirus attachment to sialic acid receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7S0P
 
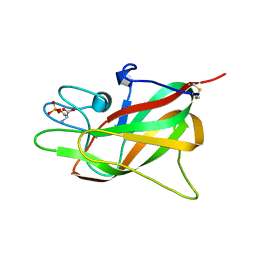 | | Crystal structure of Porcine Factor VIII C2 Domain Bound to Phosphatidylserine | | Descriptor: | Coagulation factor VIII, PHOSPHOSERINE | | Authors: | Peters, S.C, Childers, K.C, Wo, S.W, Brison, C.M, Swanson, C.D, Spiegel, P.C. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Stable binding to phosphatidylserine-containing membranes requires conserved arginine residues in tandem C domains of blood coagulation factor VIII.
Front Mol Biosci, 9, 2022
|
|
2P6Y
 
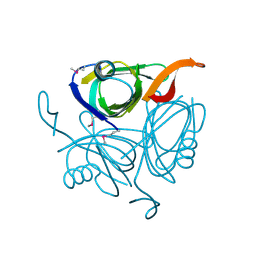 | | X-ray structure of the protein Q9KM02_VIBCH from Vibrio cholerae at the resolution 1.63 A. Northeast Structural Genomics Consortium target VcR80. | | Descriptor: | Hypothetical protein VCA0587, ZINC ION | | Authors: | Kuzin, A.P, Abashidze, M, Jayaraman, S, Chen, C.X, Wang, C, Fang, Y, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-19 | | Release date: | 2007-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray structure of the protein Q9KM02_VIBCH from Vibrio cholerae at the resolution 1.63 A.
To be Published
|
|
5XE0
 
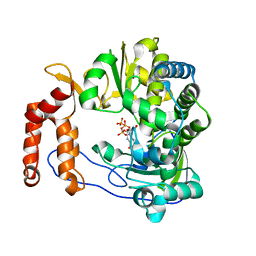 | | Crystal structure of EV-D68-3Dpol in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Genome polyprotein | | Authors: | Xie, W, Wang, C, Wang, Z, Li, Q, Wang, C. | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure and Thermostability Characterization of Enterovirus D68 3Dpol
J. Virol., 91, 2017
|
|
6XQN
 
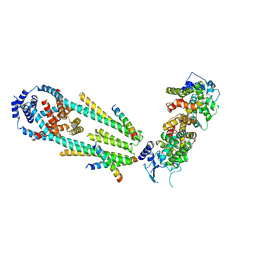 | | Structure of a mitochondrial calcium uniporter holocomplex (MICU1, MICU2, MCU, EMRE) in low Ca2+ | | Descriptor: | CALCIUM ION, Calcium uniporter protein, Calcium uptake protein 1, ... | | Authors: | Long, S.B, Wang, C, Baradaran, R, Jacewicz, A, Delgado, B. | | Deposit date: | 2020-07-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures reveal gatekeeping of the mitochondrial Ca 2+ uniporter by MICU1-MICU2.
Elife, 9, 2020
|
|
6XQO
 
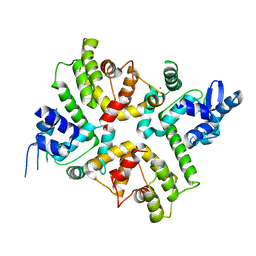 | | Structure of the human MICU1-MICU2 heterodimer, calcium bound, in association with a lipid nanodisc | | Descriptor: | CALCIUM ION, Calcium uptake protein 1, mitochondrial, ... | | Authors: | Long, S.B, Wang, C, Baradaran, R, Jacewicz, A, Delgado, B. | | Deposit date: | 2020-07-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures reveal gatekeeping of the mitochondrial Ca 2+ uniporter by MICU1-MICU2.
Elife, 9, 2020
|
|
5WZK
 
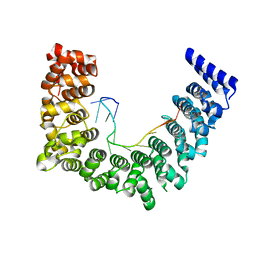 | | Structure of APUM23-deletion-of-insert-region-GGAAUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
8IF5
 
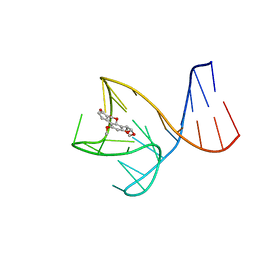 | | AFB1-AF26 APTAMER COMPLEX | | Descriptor: | AFB1 DNA aptamer (26-MER), AFLATOXIN B1 | | Authors: | Xu, G.H, Wang, C, Li, C.G. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer.
Nucleic Acids Res., 51, 2023
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
8Q5Z
 
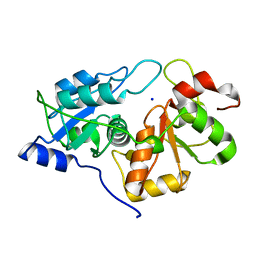 | |
6KZ6
 
 | | Crystal structure of ASFV dUTPase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R, MAGNESIUM ION | | Authors: | Guo, Y, Chen, C, Li, G.B, Cao, L, Wang, C.W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural Insight into African Swine Fever Virus dUTPase Reveals a Novel Folding Pattern in the dUTPase Family.
J.Virol., 94, 2020
|
|
5WWX
 
 | | Crystal structure of the KH2 domain of human RNA-binding E3 ubiquitin-protein ligase MEX-3C complex with RNA | | Descriptor: | NICKEL (II) ION, RNA (5'-R(P*AP*GP*AP*GP*U)-3'), RNA-binding E3 ubiquitin-protein ligase MEX3C | | Authors: | Yang, L, Wang, C, Li, F, Gong, Q. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity
J. Biol. Chem., 292, 2017
|
|
4N40
 
 | | Crystal structure of human Epithelial cell-transforming sequence 2 protein | | Descriptor: | Protein ECT2 | | Authors: | Zou, Y, Shao, Z.H, Li, F.D, Gong, D, Wang, C, Gong, Q, Shi, Y. | | Deposit date: | 2013-10-08 | | Release date: | 2014-08-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Crystal structure of triple-BRCT-domain of ECT2 and insights into the binding characteristics to CYK-4
Febs Lett., 588, 2014
|
|
4V3E
 
 | | The CIDRa domain from IT4var07 PfEMP1 bound to endothelial protein C receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOTHELIAL PROTEIN C RECEPTOR, ... | | Authors: | Lau, C.K.Y, Turner, L, Jespersen, J.S, Lowe, E.D, Petersen, B, Wang, C.W, Petersen, J.E.V, Lusingu, J, Theander, T.G, Lavstsen, T, Higgins, M.K. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Conservation Despite Huge Sequence Diversity Allows Epcr Binding by the Pfemp1 Family Implicated in Severe Childhood Malaria.
Cell Host Microbe., 17, 2015
|
|
6LOI
 
 | |
