6XRM
 
 | | Crystal structure of human PI3K-gamma in complex with Compound 4 | | Descriptor: | 5-[2-amino-3-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidin-5-yl]-2-[(1S)-1-cyclopropylethyl]-7-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Walker, N.P, Jeffrey, J.L. | | Deposit date: | 2020-07-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Discovery of Potent and Selective PI3K gamma Inhibitors.
J.Med.Chem., 63, 2020
|
|
6XRL
 
 | | Crystal structure of human PI3K-gamma in complex with inhibitor IPI-549 | | Descriptor: | 2-amino-N-[(1S)-1-{8-[(1-methyl-1H-pyrazol-4-yl)ethynyl]-1-oxo-2-phenyl-1,2-dihydroisoquinolin-3-yl}ethyl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Walker, N.P, Jeffrey, J.L. | | Deposit date: | 2020-07-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery of Potent and Selective PI3K gamma Inhibitors.
J.Med.Chem., 63, 2020
|
|
6XRN
 
 | | Crystal structure of human PI3K-gamma in complex with Compound 17 | | Descriptor: | 2-amino-5-{2-[(1S)-1-cyclopropylethyl]-7-methyl-1-oxo-2,3-dihydro-1H-isoindol-5-yl}-N-(trans-3-hydroxycyclobutyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Walker, N.P, Jeffrey, J.L. | | Deposit date: | 2020-07-13 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationship Optimization of Pyrazolopyrimidine Amide Inhibitors of Phosphoinositide 3-Kinase gamma (PI3K gamma ).
J.Med.Chem., 65, 2022
|
|
3VZD
 
 | | Crystal structure of Sphingosine Kinase 1 with inhibitor and ADP | | Descriptor: | 4-{[4-(4-chlorophenyl)-1,3-thiazol-2-yl]amino}phenol, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-10-11 | | Release date: | 2013-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of sphingosine kinase 1 substrate recognition and catalysis.
Structure, 21, 2013
|
|
3D5Q
 
 | | Crystal Structure of 11b-HSD1 in Complex with Triazole Inhibitor | | Descriptor: | 3-[1-(4-fluorophenyl)cyclopropyl]-4-(1-methylethyl)-5-[4-(trifluoromethoxy)phenyl]-4H-1,2,4-triazole, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Liu, J, Sudom, A, Walker, N.P.C. | | Deposit date: | 2008-05-16 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Distinctive molecular inhibition mechanisms for selective inhibitors of human 11beta-hydroxysteroid dehydrogenase type 1.
Bioorg.Med.Chem., 16, 2008
|
|
4XX1
 
 | | Low resolution structure of LCAT in complex with Fab1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1 heavy chain, Fab1 light chain, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T. | | Deposit date: | 2015-01-29 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The high-resolution crystal structure of human LCAT.
J.Lipid Res., 56, 2015
|
|
4XWG
 
 | | Crystal Structure of LCAT (C31Y) in complex with Fab1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1 Heavy Chain, Fab1 Light Chain, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T. | | Deposit date: | 2015-01-28 | | Release date: | 2015-07-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The high-resolution crystal structure of human LCAT.
J.Lipid Res., 56, 2015
|
|
1OVL
 
 | | Crystal Structure of Nurr1 LBD | | Descriptor: | BROMIDE ION, IODIDE ION, Orphan nuclear receptor NURR1 (MSE 414, ... | | Authors: | Wang, Z, Liu, J, Walker, N. | | Deposit date: | 2003-03-26 | | Release date: | 2003-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Function of Nurr1 identifies a Class of Ligand-Independent Nuclear Receptors
Nature, 423, 2003
|
|
3PGK
 
 | | The structure of yeast phosphoglycerate kinase at 0.25 nm resolution | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shaw, P.J, Walker, N.P, Watson, H.C. | | Deposit date: | 1982-07-15 | | Release date: | 1982-09-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence and structure of yeast phosphoglycerate kinase.
Embo J., 1, 1982
|
|
5BV7
 
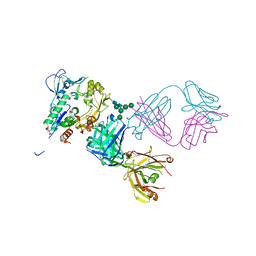 | | Crystal structure of human LCAT (L4F, N5D) in complex with Fab of an agonistic antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 27C3 heavy chain, 27C3 light chain, ... | | Authors: | Piper, D.E, Romanow, W.G, Thibault, S.T, Walker, N.P.C. | | Deposit date: | 2015-06-04 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Agonistic Human Antibodies Binding to Lecithin-Cholesterol Acyltransferase Modulate High Density Lipoprotein Metabolism.
J.Biol.Chem., 291, 2016
|
|
4L02
 
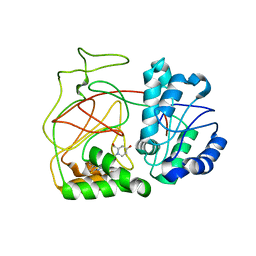 | | Crystal Structure of SphK1 with inhibitor | | Descriptor: | (2R,4S)-1-[2-(4-{[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]amino}phenyl)ethyl]-2-(hydroxymethyl)piperidin-4-ol, Sphingosine kinase 1 | | Authors: | Min, X, Walker, N, Wang, Z. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure guided design of a series of sphingosine kinase (SphK) inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4O9S
 
 | | Crystal structure of Retinol-Binding Protein 4 (RBP4)in complex with a non-retinoid ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(12),2(6),8,10-tetraen-12-yl)piperazin-1-yl]-2-[2-(trifluoromethyl)phenyl]ethanone, CHLORIDE ION, ... | | Authors: | Wang, Z, Johnstone, S, Walker, N.P. | | Deposit date: | 2014-01-02 | | Release date: | 2014-07-02 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-assisted discovery of the first non-retinoid ligands for Retinol-Binding Protein 4.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3PDJ
 
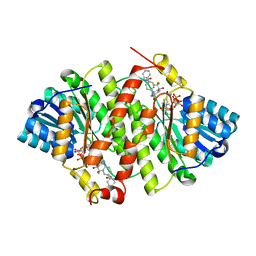 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase 1 (11b-HSD1) in Complex with 4,4-Disubstituted Cyclohexylbenzamide Inhibitor | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, N-[trans-4-(3-amino-3-oxopropyl)-4-phenylcyclohexyl]-N-cyclopropyl-4-[(1S)-2,2,2-trifluoro-1-hydroxy-1-methylethyl]benzamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and Optimization of Novel 4,4-Disubstituted Cyclohexylbenzamide Derivatives as Potent 11beta-HSD1 Inhibitors
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3OQ1
 
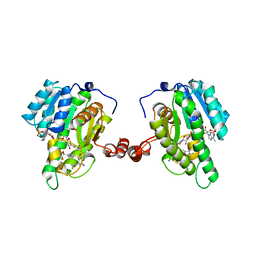 | | Crystal Structure of 11beta-Hydroxysteroid Dehydrogenase-1 (11b-HSD1) in Complex with Diarylsulfone Inhibitor | | Descriptor: | 3-(2-fluoroethyl)-4-({4-[(2S)-1,1,1-trifluoro-2-hydroxypropan-2-yl]phenyl}sulfonyl)benzonitrile, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2010-09-02 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The synthesis and SAR of novel diarylsulfone 11beta-HSD1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3QJ8
 
 | |
3D3E
 
 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) in Complex with Benzamide Inhibitor | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, N-cyclopropyl-N-(trans-4-pyridin-3-ylcyclohexyl)-4-[(1S)-2,2,2-trifluoro-1-hydroxy-1-methylethyl]benzamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Sudom, A, Liu, J, Walker, N.P. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Novel, Potent Benzamide Inhibitors of 11beta-Hydroxysteroid Dehydrogenase Type 1 (11beta-HSD1) Exhibiting Oral Activity in an Enzyme Inhibition ex Vivo Model
J.Med.Chem., 51, 2008
|
|
3VZB
 
 | | Crystal structure of Sphingosine Kinase 1 | | Descriptor: | (2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of sphingosine kinase 1 substrate recognition and catalysis.
Structure, 21, 2013
|
|
3VZC
 
 | | Crystal structure of Sphingosine Kinase 1 with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-(4-chlorophenyl)-1,3-thiazol-2-yl]amino}phenol, Sphingosine kinase 1 | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-10-11 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of sphingosine kinase 1 substrate recognition and catalysis.
Structure, 21, 2013
|
|
5C01
 
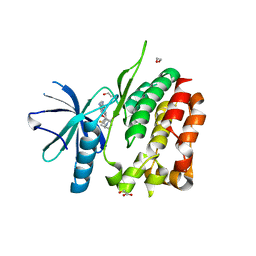 | | Crystal Structure of kinase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NON-RECEPTOR TYROSINE-PROTEIN KINASE TYK2, ... | | Authors: | Min, X, Wang, Z, Walker, N. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-16 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of the JH2 Pseudokinase Domain of JAK Family Tyrosine Kinase 2 (TYK2).
J.Biol.Chem., 290, 2015
|
|
5C03
 
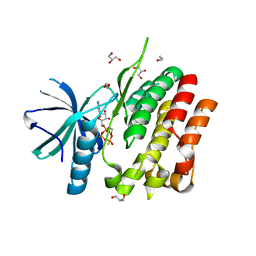 | | Crystal Structure of kinase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Min, X, Wang, Z, Walker, N. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the JH2 Pseudokinase Domain of JAK Family Tyrosine Kinase 2 (TYK2).
J.Biol.Chem., 290, 2015
|
|
1Z8G
 
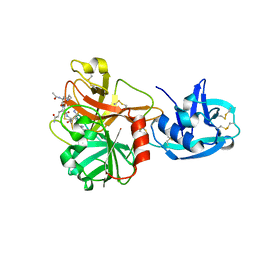 | | Crystal structure of the extracellular region of the transmembrane serine protease hepsin with covalently bound preferred substrate. | | Descriptor: | ACE-LYS-GLN-LEU-ARG-Chloromethylketone, Serine protease hepsin | | Authors: | Herter, S, Piper, D.E, Aaron, W, Gabriele, T, Cutler, G, Cao, P, Bhatt, A.S, Choe, Y, Craik, C.S, Walker, N, Meininger, D, Hoey, T, Austin, R.J. | | Deposit date: | 2005-03-30 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Hepatocyte growth factor is a preferred in vitro substrate for human hepsin, a membrane-anchored serine protease implicated in prostate and ovarian cancers
Biochem.J., 390, 2005
|
|
3QKV
 
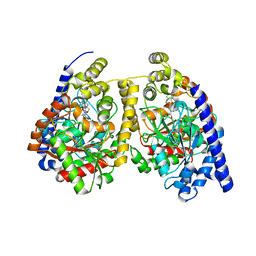 | | Crystal structure of fatty acid amide hydrolase with small molecule compound | | Descriptor: | (6-bromo-1'H,4H-spiro[1,3-benzodioxine-2,4'-piperidin]-1'-yl)methanol, Fatty-acid amide hydrolase 1 | | Authors: | Min, X, Walker, N.P.C, Wang, Z. | | Deposit date: | 2011-02-01 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QJ9
 
 | | Crystal structure of fatty acid amide hydrolase with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-{(3S)-1-[4-(1-benzofuran-2-yl)pyrimidin-2-yl]piperidin-3-yl}-3-ethyl-1,3-dihydro-2H-benzimidazol-2-one, Fatty-acid amide hydrolase 1, ... | | Authors: | Min, X, Walker, N.P.C, Wang, Z. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3CZR
 
 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) in Complex with Arylsulfonylpiperazine Inhibitor | | Descriptor: | (2R)-1-[(4-tert-butylphenyl)sulfonyl]-2-methyl-4-(4-nitrophenyl)piperazine, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Corticosteroid 11-beta-dehydrogenase isozyme 1, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2008-04-29 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and Initial SAR of Arylsulfonylpiperazine Inhibitors of 11beta-Hydroxysteroid Dehydrogenase Type 1 (11beta-HSD1)
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3D4N
 
 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) in Complex with Sulfonamide Inhibitor | | Descriptor: | 1-{[(3R)-3-methyl-4-({4-[(1S)-2,2,2-trifluoro-1-hydroxy-1-methylethyl]phenyl}sulfonyl)piperazin-1-yl]methyl}cyclopropanecarboxamide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Liu, J, Sudom, A, Walker, N.P. | | Deposit date: | 2008-05-14 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Novel, Potent Benzamide Inhibitors of 11beta-Hydroxysteroid Dehydrogenase Type 1 (11beta-HSD1) Exhibiting Oral Activity in an Enzyme Inhibition ex Vivo Model
J.Med.Chem., 51, 2008
|
|
