6U60
 
 | | Crystal structure of prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD and L-tyrosine | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Prephenate dehydrogenase, ... | | 著者 | Shabalin, I.G, Hou, J, Kutner, J, Grimshaw, S, Christendat, D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-08-28 | | 公開日 | 2019-09-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
6U6J
 
 | | RNA-monomer complex containing pyrophosphate linkage | | 分子名称: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(hydroxy)phosphoryl]guanosine, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(DPG))-3') | | 著者 | Zhang, W, Szostak, J.W, Giurgiu, C, Wright, T. | | 登録日 | 2019-08-29 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Prebiotically Plausible "Patching" of RNA Backbone Cleavage through a 3'-5' Pyrophosphate Linkage.
J.Am.Chem.Soc., 141, 2019
|
|
3DHC
 
 | | 1.3 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homocysteine Bound to The catalytic Metal Center | | 分子名称: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homocysteine, ... | | 著者 | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | 登録日 | 2008-06-17 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
6U8F
 
 | | RNA hairpin, bound with TNA monomer | | 分子名称: | 2-azanyl-9-[(2~{R},3~{R},4~{S})-3-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-oxolan-2-yl]-1~{H}-purin-6-one, RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*CP*AP*UP*GP*AP*AP*AP*GP*U)-3'), RNA (5'-R(P*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3') | | 著者 | Szostak, J.W, Zhang, W. | | 登録日 | 2019-09-04 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
6UE5
 
 | | Crystal structure of full-length human DCAF15-DDB1-deltaPBP-DDA1-RBM39 in complex with 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzenesulfonamide | | 分子名称: | 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | 著者 | Knapp, M.S, Shu, W, Xie, L, Bussiere, D.E. | | 登録日 | 2019-09-20 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
6U5G
 
 | | MicroED structure of a FIB-milled CypA Crystal | | 分子名称: | Peptidyl-prolyl cis-trans isomerase A | | 著者 | Wolff, A.M, Martynowycz, M.W, Zhao, W, Gonen, T, Fraser, J.S, Thompson, M.C. | | 登録日 | 2019-08-27 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | 主引用文献 | Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals
Iucrj, 7, 2020
|
|
2YJ1
 
 | | Puma BH3 foldamer in complex with Bcl-xL | | 分子名称: | ALPHA-BETA-PUMA BH3 FOLDAMER, BCL-2-LIKE PROTEIN 1 | | 著者 | Lee, E.F, Smith, B.J, Horne, W.S, Mayer, K.N, Evangelista, M, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | 登録日 | 2011-05-18 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural Basis of Bcl-Xl Recognition by a Bh3-Mimetic Alpha-Beta-Peptide Generated Via Sequence-Based Design
Chembiochem, 12, 2011
|
|
1BS8
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | 分子名称: | PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
6U7Y
 
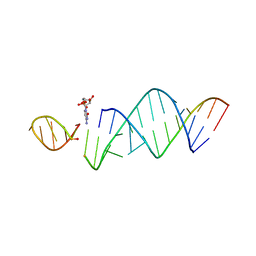 | | RNA hairpin structure containing one TNA nucleotide as template | | 分子名称: | CYTIDINE-5'-MONOPHOSPHATE, RNA (5'-R(*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3'), RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*(TG)P*CP*CP*GP*AP*AP*AP*GP*G)-3') | | 著者 | Szostak, J.W, Zhang, W. | | 登録日 | 2019-09-03 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
1BS5
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM | | 分子名称: | PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ZINC ION | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
6OEM
 
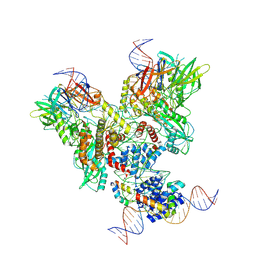 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA0) | | 分子名称: | DNA (46-MER), DNA (57-MER), High mobility group protein B1, ... | | 著者 | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | 登録日 | 2019-03-27 | | 公開日 | 2020-01-29 | | 最終更新日 | 2020-02-26 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6TUM
 
 | |
1QFR
 
 | | NMR SOLUTION STRUCTURE OF PHOSPHOCARRIER PROTEIN HPR FROM ENTEROCOCCUS FAECALIS | | 分子名称: | PHOSPHOCARRIER PROTEIN HPR | | 著者 | Maurer, T, Doeker, R, Goerler, A, Hengstenberg, W, Kalbitzer, H.R. | | 登録日 | 1999-04-13 | | 公開日 | 2001-02-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of the histidine-containing phosphocarrier protein (HPr) from Enterococcus faecalis in solution.
Eur.J.Biochem., 268, 2001
|
|
6UKT
 
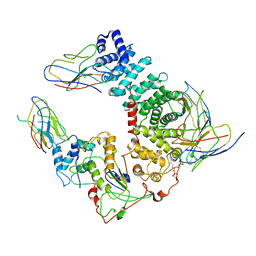 | | Cryo-EM structure of mammalian Ric-8A:Galpha(i):nanobody complex | | 分子名称: | Guanine nucleotide-binding protein G(i) subunit alpha-1, NB8109, NB8117, ... | | 著者 | Mou, T.C, Zhang, K, Johnston, J.D, Chiu, W, Sprang, S.R. | | 登録日 | 2019-10-05 | | 公開日 | 2020-03-11 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | Structure of the G protein chaperone and guanine nucleotide exchange factor Ric-8A bound to G alpha i1.
Nat Commun, 11, 2020
|
|
6U9I
 
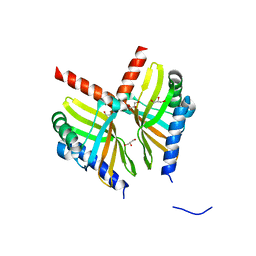 | | Crystal structure of BvnE pinacolase from Penicillium brevicompactum | | 分子名称: | BvnE, DI(HYDROXYETHYL)ETHER, GLYCEROL | | 著者 | Ye, Y, Du, L, Zhang, X, Newmister, S.A, McCauley, M, Alegre-Requena, J.V, Zhang, W, Mu, S, Minami, A, Fraley, A.E, Adrover-Castellano, M.L, Carney, N, Shende, V.V, Oikawa, H, Kato, H, Tsukamoto, S, Paton, R.S, Williams, R.M, Sherman, D.H, Li, S. | | 登録日 | 2019-09-09 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.777 Å) | | 主引用文献 | Fungal-derived brevianamide assembly by a stereoselective semipinacolase.
Nat Catal, 3, 2020
|
|
6OEP
 
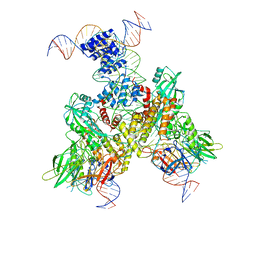 | | Cryo-EM structure of mouse RAG1/2 12RSS-NFC/23RSS-PRC complex (DNA1) | | 分子名称: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | 著者 | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | 登録日 | 2019-03-27 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UBL
 
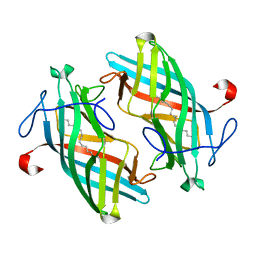 | | Structure of DynF from the Dynemicin Biosynthesis Pathway of Micromonospora chersina | | 分子名称: | DynF, PALMITIC ACID | | 著者 | Kosgei, A.J, Miller, M.D, Xu, W, Bhardwaj, M, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | 登録日 | 2019-09-12 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6OGK
 
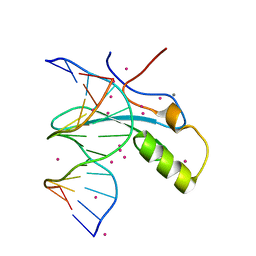 | | MeCP2 MBD in complex with DNA | | 分子名称: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*CP*TP*CP*CP*G)-3'), ... | | 著者 | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | 登録日 | 2019-04-02 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
6UGH
 
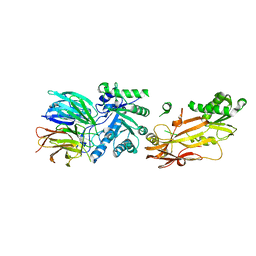 | |
6OEN
 
 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA1) | | 分子名称: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | 著者 | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | 登録日 | 2019-03-27 | | 公開日 | 2020-01-29 | | 最終更新日 | 2020-02-26 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OFC
 
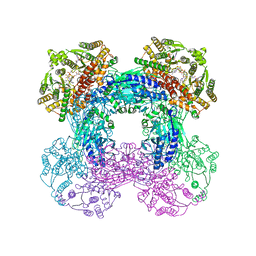 | | Crystal structure of M. tuberculosis glutamine-dependent NAD+ synthetase complexed with Sulfonamide derivative 1, pyrophosphate, and glutamine | | 分子名称: | 5'-O-[(pyridine-3-carbonyl)sulfamoyl]adenosine, CHLORIDE ION, GLUTAMINE, ... | | 著者 | Chuenchor, W, Doukov, T.I, Gerratana, B. | | 登録日 | 2019-03-28 | | 公開日 | 2020-01-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Different ways to transport ammonia in human and Mycobacterium tuberculosis NAD+synthetases.
Nat Commun, 11, 2020
|
|
6OM7
 
 | |
6U5A
 
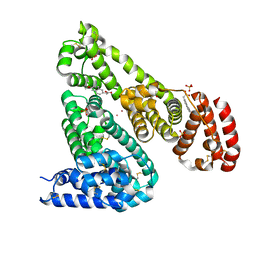 | | Crystal structure of Equine Serum Albumin complex with 6-MNA | | 分子名称: | (6-methoxynaphthalen-2-yl)acetic acid, SULFATE ION, Serum albumin, ... | | 著者 | Czub, M.P, Handing, K.B, Venkataramany, B.S, Cymborowski, M.T, Shabalin, I.G, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-08-27 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Albumin-Based Transport of Nonsteroidal Anti-Inflammatory Drugs in Mammalian Blood Plasma.
J.Med.Chem., 63, 2020
|
|
6OLZ
 
 | |
6U5Z
 
 | |
