5BP7
 
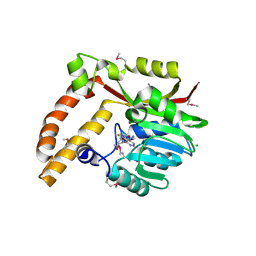 | | Crystal structure of SAM-dependent methyltransferase from Geobacter sulfurreducens in complex with S-Adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Kutner, J, Shabalin, I.G, Mason, D.V, Handing, K.B, Gasiorowska, O.A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Geobacter sulfurreducens in complex with S-Adenosyl-L-homocysteine
to be published
|
|
5J23
 
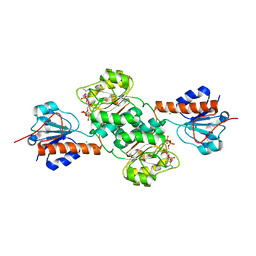 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with 2'-phospho-ADP-ribose | | Descriptor: | 2-hydroxyacid dehydrogenase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Shabalin, I.G, Gasiorowska, O.A, Handing, K.B, Bonanno, J, Kutner, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-29 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5EPE
 
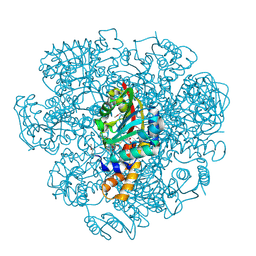 | | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase, SODIUM ION | | Authors: | LaRowe, C, Shabalin, I.G, Kutner, J, Handing, K.B, Stead, M, Hillerich, B.S, Ahmed, M, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-11-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine
to be published
|
|
6U60
 
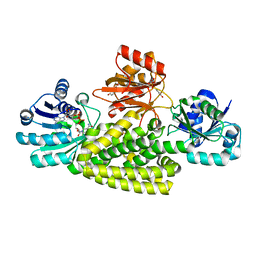 | | Crystal structure of prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD and L-tyrosine | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Prephenate dehydrogenase, ... | | Authors: | Shabalin, I.G, Hou, J, Kutner, J, Grimshaw, S, Christendat, D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-28 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
5TX7
 
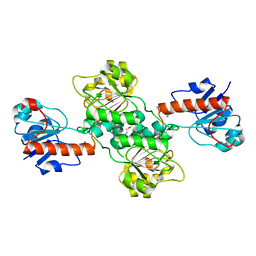 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Desulfovibrio vulgaris | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase family protein, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Czub, M.P, Shabalin, I.G, Gasiorowska, O.A, Handing, K.B, Kutner, J, Cymborowski, M.T, Hennig, P.M, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-11-15 | | Release date: | 2016-12-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Desulfovibrio vulgaris
to be published
|
|
5V7N
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADP and 2-Keto-D-gluconic acid | | Descriptor: | 2-keto-D-gluconic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Handing, K.B, Miks, C.D, Kutner, J, Matelska, D, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5V7G
 
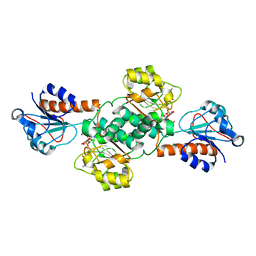 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADPH and oxalate | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shabalin, I.G, Mason, D.V, Handing, K.B, Kutner, J, Matelska, D, Cooper, D.R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5UNN
 
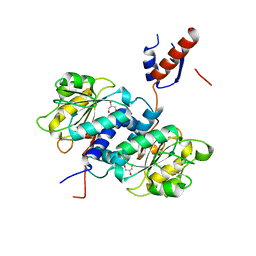 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in apo form | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH-dependent glyoxylate/hydroxypyruvate reductase | | Authors: | Shabalin, I.G, LaRowe, C, Kutner, J, Gasiorowska, O.A, Handing, K.B, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-01-31 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5TSD
 
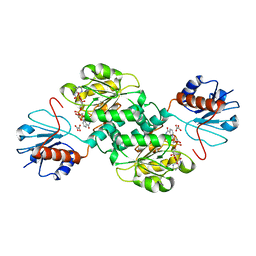 | | Crystal structure of NADPH-dependent 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH and oxalate | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OXALIC ACID, Probable hydroxyacid dehydrogenase protein | | Authors: | Matelska, D, Shabalin, I.G, Kutner, J, Handing, K.B, Gasiorowska, O.A, Cooper, D.R, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-10-28 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of NADPH-dependent 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH and oxalate
to be published
|
|
5VG6
 
 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 in complex with NADPH and MES. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lipowska, J, Shabalin, I.G, Kutner, J, Gasiorowska, O.A, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-04-10 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 in complex with NADPH and MES.
to be published
|
|
