4COO
 
 | | Crystal structure of human cystathionine beta-synthase (delta516-525) at 2.0 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETATE ION, ... | | Authors: | McCorvie, T.J, Kopec, J, Vollamar, M, Strain-Damerell, C, Bushell, S, Bradley, A, Tallant, C, Kiyani, W, Froese, D.S, Carpenter, E.S, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-01-29 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inter-Domain Communication of Human Cystathionine Beta Synthase: Structural Basis of S-Adenosyl-L-Methionine Activation.
J.Biol.Chem., 289, 2014
|
|
4JLR
 
 | | Crystal structure of a designed Respiratory Syncytial Virus Immunogen in complex with Motavizumab | | Descriptor: | Motavizumab Fab heavy chain, Motavizumab Fab light chain, PENTAETHYLENE GLYCOL, ... | | Authors: | Rupert, P.B, Correia, B, Schief, W, Strong, R.K. | | Deposit date: | 2013-03-12 | | Release date: | 2014-02-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Proof of principle for epitope-focused vaccine design.
Nature, 507, 2014
|
|
4JJ9
 
 | | Crystal Structure of 5-carboxymethyl-2-hydroxymuconate delta-isomerase | | Descriptor: | 5-carboxymethyl-2-hydroxymuconate delta-isomerase, SULFATE ION | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-07 | | Release date: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of 5-carboxymethyl-2-hydroxymuconate delta-isomerase
To be Published
|
|
4NXO
 
 | | Crystal Structure of Insulin Degrading Enzyme in complex with BDM44768 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Deprez, B, Tang, W. | | Deposit date: | 2013-12-09 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
4FCG
 
 | | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Zimmerman, M.D, Minor, W, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-13 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL)
To be Published
|
|
4NW3
 
 | | Crystal structure of MLL CXXC domain in complex with a CpG DNA | | Descriptor: | 5'-D(*GP*CP*CP*AP*TP*CP*GP*AP*TP*GP*GP*C)-3', Histone-lysine N-methyltransferase 2A, ZINC ION | | Authors: | Bian, C, Tempel, W, Chao, X, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-05 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
4O64
 
 | | Zinc fingers of KDM2B | | Descriptor: | Lysine-specific demethylase 2B, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Liu, K, Xu, C, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
4F3S
 
 | | Crystal structure of periplasmic D-alanine ABC transporter from Salmonella enterica | | Descriptor: | D-ALANINE, GLYCINE, PHOSPHATE ION, ... | | Authors: | Agarwal, R, Chamala, S, Evans, B, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Foti, R, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of periplasmic D-alanine ABC transporter from Salmonella enterica
To be Published
|
|
8GXR
 
 | |
8G6U
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with 8ANC195 and 10-1074 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CRF-1_AE T/F100 HIV-1 gp41, ... | | Authors: | Chen, Y, Zhou, F, Huang, R, Tolbert, W, Pazgier, M. | | Deposit date: | 2023-02-16 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
8H70
 
 | |
3RPL
 
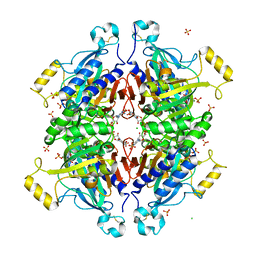 | | D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase of Synechocystis sp. PCC 6803 in complex with FRUCTOSE-1,6-BISPHOSPHATE | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Hu, X, Hui, D, Lingling, F, Jian, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New insights into the structural and interactional basis for a promising route towards fructose-1,6-/sedoheptulose-1,7-bisphosphatases controlling
To be Published
|
|
3SMR
 
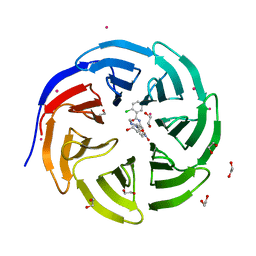 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Wasney, G.A, Tempel, W, Senisterra, G, Bolshan, Y, Smil, D, Nguyen, K.T, Hajian, T, Poda, G, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
3SSZ
 
 | | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Rhodobacteraceae bacterium | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme, N-terminal domain protein, SULFATE ION | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Rhodobacteraceae bacterium
To be Published
|
|
8IB0
 
 | |
3T8Q
 
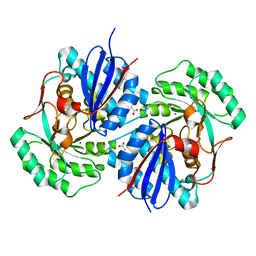 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme family protein from Hoeflea phototrophica | | Descriptor: | MAGNESIUM ION, MALONATE ION, Mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme family protein from Hoeflea phototrophica
To be Published
|
|
8IKJ
 
 | | Cryo-EM structure of the inactive CD97 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor E5,Soluble cytochrome b562,Adhesion G protein-coupled receptor E5 subunit beta | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P, Guo, J, Qin, J, Shen, D, Ji, S, Zang, S, Zhang, H, Wang, W, Shen, Q, Sun, P, Zhang, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
3T1Y
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA Lysine 3 (TRNALYS3) bound to an mRNA with an AAG-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
3RQ4
 
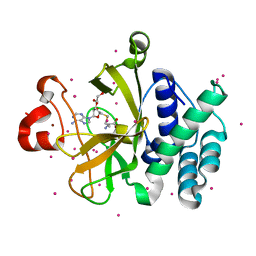 | | Crystal structure of suppressor of variegation 4-20 homolog 2 | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SUV420H2, S-ADENOSYLMETHIONINE, ... | | Authors: | Dong, A, Zeng, H, Tempel, W, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-27 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2.
Febs Lett., 587, 2013
|
|
8I13
 
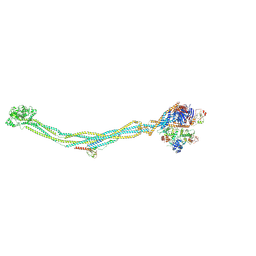 | | Cryo-EM structure of 6-subunit Smc5/6 | | Descriptor: | MMS21 isoform 1, NSE3 isoform 1, Non-structural maintenance of chromosomes element 1 homolog, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4V
 
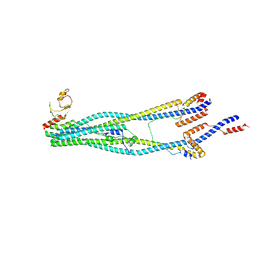 | | Cryo-EM structure of 5-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.97 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4W
 
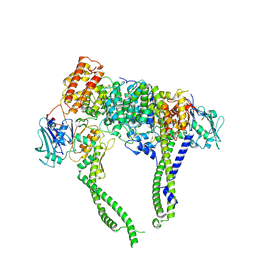 | | Cryo-EM structure of 5-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Cheng, T, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.01 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4X
 
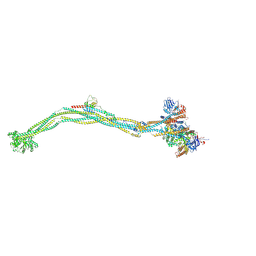 | | Cryo-EM structure of 5-subunit Smc5/6 | | Descriptor: | E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 5, Nse6, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
3T61
 
 | | Crystal Structure of a gluconokinase from Sinorhizobium meliloti 1021 | | Descriptor: | Gluconokinase, PHOSPHATE ION | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-28 | | Release date: | 2011-08-17 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a gluconokinase from Sinorhizobium meliloti 1021
To be Published
|
|
3T4W
 
 | | The crystal structure of mandelate racemase/muconate lactonizing enzyme from Sulfitobacter sp | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-26 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | The crystal structure of mandelate racemase/muconate lactonizing enzyme from Sulfitobacter sp
To be Published
|
|
