6XOI
 
 | | Structure of SUMO1-ML00752641 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XDB
 
 | | Crystal structure of IRE1a in complex with G-6904 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-N-(6-chloro-2-fluoro-3-{[(2-fluorophenyl)sulfonyl]amino}phenyl)-6-(1,3-dimethyl-1H-pyrazol-4-yl)quinazoline-8-carboxamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H.A, Weiru, W. | | Deposit date: | 2020-06-10 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of BRaf-Sparing Amino-Thienopyrimidines with Potent IRE1 alpha Inhibitory Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
2LGL
 
 | | NMR structure of the UHRF1 PHD domain | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
3B9O
 
 | | long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN | | Descriptor: | Alkane monooxygenase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
6Y1A
 
 | | Amyloid fibril structure of islet amyloid polypeptide | | Descriptor: | AMINO GROUP, Islet amyloid polypeptide | | Authors: | Roeder, C, Kupreichyk, T, Gremer, L, Schaefer, L.U, Pothula, K.R, Ravelli, R.B.G, Willbold, D, Hoyer, W, Schroder, G.F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-03-04 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of islet amyloid polypeptide fibrils reveals similarities with amyloid-beta fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2FDP
 
 | | Crystal structure of beta-secretase complexed with an amino-ethylene inhibitor | | Descriptor: | Beta-secretase 1, N1-((2S,3S,5R)-3-AMINO-6-(4-FLUOROPHENYLAMINO)-5-METHYL-6-OXO-1-PHENYLHEXAN-2-YL)-N3,N3-DIPROPYLISOPHTHALAMIDE | | Authors: | Yang, W, Lu, W, Lu, Y, Zhong, M, Sun, J, Thomas, A.E, Wilkinson, J.M, Fucini, R.V, Lam, M, Randal, M, Shi, X.P, Jacobs, J.W, McDowell, R.S, Gordon, E.M, Ballinger, M.D. | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminoethylenes: a tetrahedral intermediate isostere yielding potent inhibitors of the aspartyl protease BACE-1.
J.Med.Chem., 49, 2006
|
|
2M5X
 
 | | Novel method of protein purification for structural research. Example of ultra high resolution structure of SPI-2 inhibitor by X-ray and NMR spectroscopy. | | Descriptor: | Silk protease inhibitor 2 | | Authors: | Lenarcic Zivkovic, M, Dvornyk, A, Kludkiewicz, B, Kopera, E, Zagorski-Ostoja, W, Grzelak, K, Zhukov, I, Bal, W. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution structure of a protein prepared by non-enzymatic His-tag removal. Crystallographic and NMR study of GmSPI-2 inhibitor.
Plos One, 9, 2014
|
|
6WVB
 
 | | Takifugu rubripes VKOR-like with warfarin | | Descriptor: | S-WARFARIN, Vitamin K epoxide reductase-like protein, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.872 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WV6
 
 | | Human VKOR with phenindione | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phenindione, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WV3
 
 | | Human VKOR with warfarin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, S-WARFARIN, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
3HM8
 
 | | Crystal structure of the C-terminal Hexokinase domain of human HK3 | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Hexokinase-3, alpha-D-glucopyranose | | Authors: | Nedyalkova, L, Tong, Y, Rabeh, W, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the C-terminal Hexokinase domain of human HK3
To be Published
|
|
2MA1
 
 | | Solution structure of HRDC1 domain of RecQ helicase from Deinococcus radiodurans | | Descriptor: | DNA helicase RecQ | | Authors: | Liu, S, Zhang, W, Gao, Z, Ming, Q, Hou, H, Lan, W, Wu, H, Cao, C, Dong, Y. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal-most HRDC1 domain of RecQ helicase from Deinococcus radiodurans
To be Published
|
|
3B4O
 
 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, apo form | | Descriptor: | ACETATE ION, Phenazine biosynthesis protein A/B | | Authors: | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2007-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
6WVH
 
 | | Human VKOR with Brodifacoum | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Brodifacoum, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
3B9N
 
 | | Crystal structure of long-chain alkane monooxygenase (LadA) | | Descriptor: | Alkane monooxygenase | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
3MEF
 
 | |
3B4P
 
 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, complex with 2-(cyclohexylamino)benzoic acid | | Descriptor: | 2-(cyclohexylamino)benzoic acid, ACETATE ION, AZIDE ION, ... | | Authors: | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2007-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3IHI
 
 | | Crystal structure of mouse thymidylate synthase | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W.R, Sokolowska, M, Fraczyk, T, Ciesla, J, Rode, W. | | Deposit date: | 2009-07-30 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mouse thymidylate synthase does not show the inactive conformation, observed for the human enzyme
Struct Chem, 2016
|
|
8GYF
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (K192Y) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, staygold(K192Y) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2022-09-22 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (K192Y) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8H4R
 
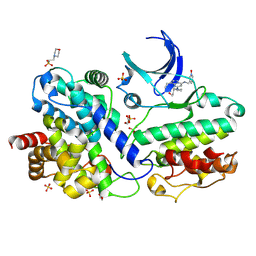 | | The Crystal Structure of CDK3 and CyclinE1 Complex with Dinaciclib from Biortus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, G1/S-specific cyclin-E1, ... | | Authors: | Gui, W, Wang, F, Cheng, W, Gao, J, Huang, Y, Ouyang, Z. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of CDK3 and CyclinE1 Complex with Dinaciclib from Biortus
To Be Published
|
|
2F5T
 
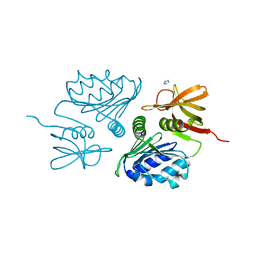 | | Crystal Structure of the sugar binding domain of the archaeal transcriptional regulator TrmB | | Descriptor: | IMIDAZOLE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, archaeal transcriptional regulator TrmB | | Authors: | Krug, M, Lee, S.J, Diederichs, K, Boos, W, Welte, W. | | Deposit date: | 2005-11-27 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of the Sugar Binding Domain of the Archaeal Transcriptional Regulator TrmB
J.Biol.Chem., 281, 2006
|
|
3CNM
 
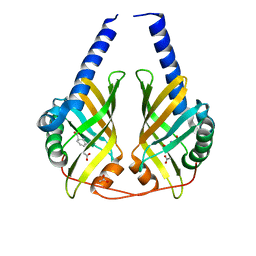 | | Crystal Structure of Phenazine Biosynthesis Protein PhzA/B from Burkholderia cepacia R18194, DHHA complex | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, ACETATE ION, Phenazine biosynthesis protein A/B | | Authors: | Ahuja, E.G, Blankenfeldt, W. | | Deposit date: | 2008-03-26 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
3IB3
 
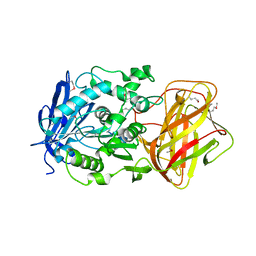 | | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Domagalski, M.J, Chruszcz, M, Osinski, T, Skarina, T, Onopriyenko, O, Cymborowski, M, Shumilin, I.A, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus
To be Published
|
|
1DP7
 
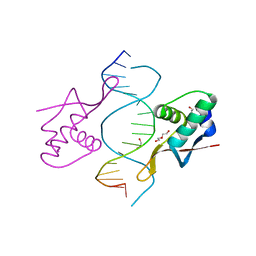 | | COCRYSTAL STRUCTURE OF RFX-DBD IN COMPLEX WITH ITS COGNATE X-BOX BINDING SITE | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*GP*(BRU)P*TP*AP*CP*CP*AP*(BRU)P*GP*GP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Gajiwala, K.S, Chen, H, Cornille, F, Roques, B.P, Reith, W, Mach, B, Burley, S.K. | | Deposit date: | 1999-12-23 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding.
Nature, 403, 2000
|
|
