6NZU
 
 | | Structure of the human frataxin-bound iron-sulfur cluster assembly complex | | Descriptor: | Acyl carrier protein, Cysteine desulfurase, mitochondrial, ... | | Authors: | Fox, N.G, Yu, X, Xidong, F, Alain, M, Joseph, N, Claire, S.D, Christine, B, Han, S, Yue, W.W. | | Deposit date: | 2019-02-14 | | Release date: | 2019-05-22 | | Last modified: | 2019-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism.
Nat Commun, 10, 2019
|
|
3P0U
 
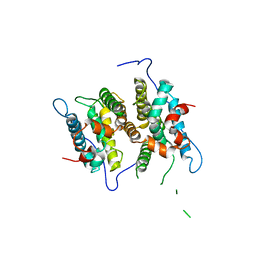 | | Crystal Structure of the ligand binding domain of human testicular receptor 4 | | Descriptor: | Nuclear receptor subfamily 2 group C member 2 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Xu, Y, Chan, C.-W, Kruse, S.W, Reynolds, R, Engel, J.D, Xu, H.E. | | Deposit date: | 2010-09-29 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Orphan Nuclear Receptor TR4 Is a Vitamin A-activated Nuclear Receptor.
J.Biol.Chem., 286, 2011
|
|
2PM1
 
 | |
1D0I
 
 | | CRYSTAL STRUCTURE OF TYPE II DEHYDROQUINASE FROM STREPTOMYCES COELICOLOR COMPLEXED WITH PHOSPHATE IONS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, TYPE II 3-DEHYDROQUINATE HYDRATASE | | Authors: | Roszak, A.W, Krell, T, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 1999-09-10 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure and mechanism of the type II dehydroquinase from Streptomyces coelicolor.
Structure, 10, 2002
|
|
1CQZ
 
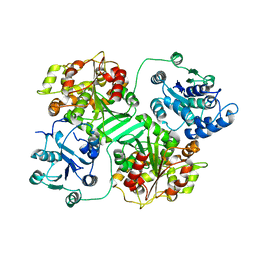 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE. | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Argiriadi, M.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7VVC
 
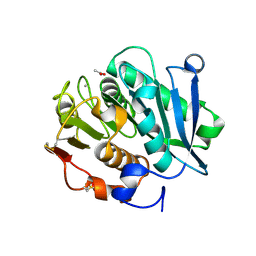 | | Crystal structure of inactive mutant of leaf-branch compost cutinase variant | | Descriptor: | ACETATE ION, ACETIC ACID, CALCIUM ION, ... | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
2GSS
 
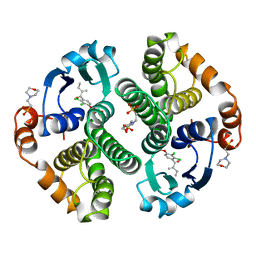 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 IN COMPLEX WITH ETHACRYNIC ACID | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ETHACRYNIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, ... | | Authors: | Oakley, A.J, Rossjohn, J, Parker, M.W. | | Deposit date: | 1996-10-29 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The three-dimensional structure of the human Pi class glutathione transferase P1-1 in complex with the inhibitor ethacrynic acid and its glutathione conjugate.
Biochemistry, 36, 1997
|
|
1CVE
 
 | |
4DHL
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment MO07123 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-methylphenyl)-1,3-thiazole-4-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Clayton, D.J, Hussein, W.M, Schenk, G, McGeary, R, Guddat, L.W. | | Deposit date: | 2012-01-29 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
6KEU
 
 | | Wildtype E53, a microbial HSL esterase | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Yang, X.C, Li, Z.Y, Xu, X.W, Li, J.X. | | Deposit date: | 2019-07-05 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Wildtype E53, a microbial HSL esterase
To Be Published
|
|
1L96
 
 | | STRUCTURE OF A HINGE-BENDING BACTERIOPHAGE T4 LYSOZYME MUTANT, ILE3-> PRO | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Dixon, M, Shewchuk, L, Matthews, B.W. | | Deposit date: | 1992-02-11 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a hinge-bending bacteriophage T4 lysozyme mutant, Ile3-->Pro.
J.Mol.Biol., 227, 1992
|
|
6NZJ
 
 | | Structural Analysis of a Nitrogenase Iron Protein from Methanosarcina acetivorans: Implications for CO2 Capture by a Surface-Exposed [Fe4S4] Cluster | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein, SULFATE ION | | Authors: | Rettberg, L.A, Kang, W, Stiebritz, M.T, Hiller, C.J, Lee, C.C, Liedtke, J, Ribbe, M.W, Hu, Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of a Nitrogenase Iron Protein from Methanosarcina acetivorans: Implications for CO 2 Capture by a Surface-Exposed [Fe 4 S 4 ] Cluster.
Mbio, 10, 2019
|
|
5V7N
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADP and 2-Keto-D-gluconic acid | | Descriptor: | 2-keto-D-gluconic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Handing, K.B, Miks, C.D, Kutner, J, Matelska, D, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
1CRC
 
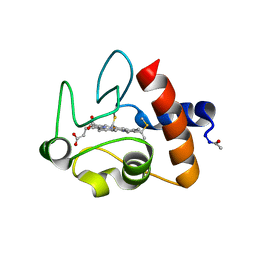 | | CYTOCHROME C AT LOW IONIC STRENGTH | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Sanishvili, R, Volz, K.W, Westbrook, E.M, Margoliash, E. | | Deposit date: | 1995-03-22 | | Release date: | 1996-03-08 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The low ionic strength crystal structure of horse cytochrome c at 2.1 A resolution and comparison with its high ionic strength counterpart.
Structure, 3, 1995
|
|
6KIG
 
 | | Structure of cyanobacterial photosystem I-IsiA supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
1CG1
 
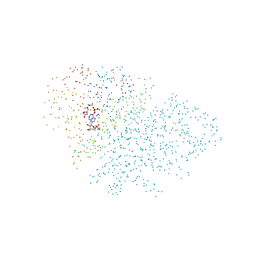 | | STRUCTURE OF THE MUTANT (K16Q) OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI COMPLEXED WITH HADACIDIN, GDP, 6-PHOSPHORYL-IMP, AND MG2+ | | Descriptor: | 6-O-PHOSPHORYL INOSINE MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, HADACIDIN, ... | | Authors: | Choe, J.Y, Poland, B.W, Fromm, H, Honzatko, R. | | Deposit date: | 1999-03-26 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic implications from crystalline complexes of wild-type and mutant adenylosuccinate synthetases from Escherichia coli.
Biochemistry, 38, 1999
|
|
2TBD
 
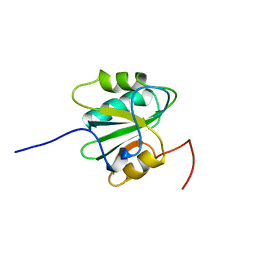 | | SV40 T ANTIGEN DNA-BINDING DOMAIN, NMR, 30 STRUCTURES | | Descriptor: | SV40 T ANTIGEN | | Authors: | Luo, X, Sanford, D.G, Bullock, P.A, Bachovchin, W.W. | | Deposit date: | 1997-01-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the origin DNA-binding domain of SV40 T-antigen.
Nat.Struct.Biol., 3, 1996
|
|
1CJB
 
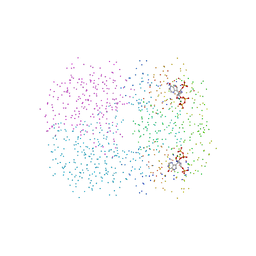 | | MALARIAL PURINE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | (1S)-1(9-DEAZAHYPOXANTHIN-9YL)1,4-DIDEOXY-1,4-IMINO-D-RIBITOL-5-PHOSPHATE, MAGNESIUM ION, PROTEIN (HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE), ... | | Authors: | Shi, W, Li, C.M, Tyler, P.C, Furneaux, R.H, Cahill, S.M, Girvin, M.E, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-04-08 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of malarial purine phosphoribosyltransferase in complex with a transition-state analogue inhibitor.
Biochemistry, 38, 1999
|
|
1RO7
 
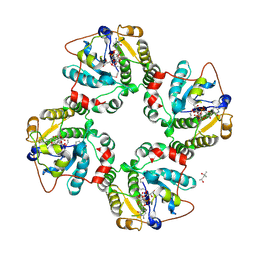 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, CMP-3FNeuAc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
5UKF
 
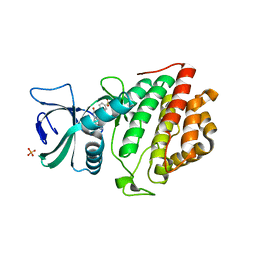 | | Crystal Structure of the Human Vaccinia-related Kinase 1 Bound to an Oxindole Inhibitor | | Descriptor: | 4-{[(Z)-(7-oxo-6,7-dihydro-8H-[1,3]thiazolo[5,4-e]indol-8-ylidene)methyl]amino}benzene-1-sulfonamide, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Counago, R.M, Wells, C, Zuercher, W, Willson, T.M, Bountra, C, Edwards, A.M, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-22 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of human Vaccinia-Related Kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations.
Sci Rep, 7, 2017
|
|
1RPR
 
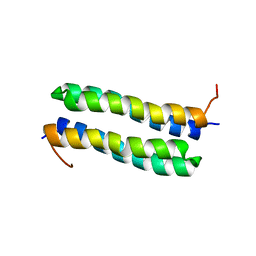 | | THE STRUCTURE OF COLE1 ROP IN SOLUTION | | Descriptor: | ROP | | Authors: | Eberle, W, Pastore, A, Klaus, W, Sander, C, Roesch, P. | | Deposit date: | 1991-10-09 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of ColE1 rop in solution.
J.Biomol.NMR, 1, 1991
|
|
1RGS
 
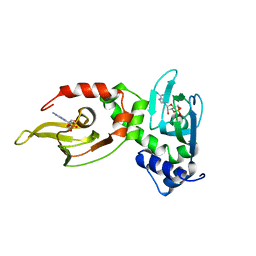 | | REGULATORY SUBUNIT OF CAMP DEPENDENT PROTEIN KINASE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP DEPENDENT PROTEIN KINASE | | Authors: | Su, Y, Dostmann, W.R.G, Herberg, F.W, Durick, K, Xuong, N.-H, Ten Eyck, L, Taylor, S.S, Varughese, K.I. | | Deposit date: | 1995-06-21 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains.
Science, 269, 1995
|
|
3AZY
 
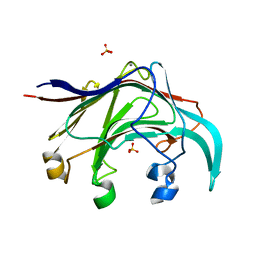 | | Crystal structure of the laminarinase catalytic domain from Thermotoga maritima MSB8 | | Descriptor: | CALCIUM ION, Laminarinase, SULFATE ION | | Authors: | Jeng, W.Y, Wang, N.C, Wang, A.H.J. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with inhibitors: essential residues for beta-1,3 and beta-1,4 glucan selection.
J.Biol.Chem., 286, 2011
|
|
2UXE
 
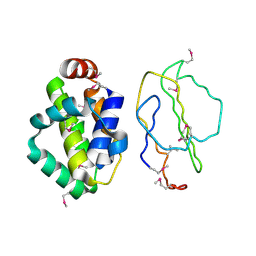 | | The structure of Vaccinia virus N1 | | Descriptor: | HYPOTHETICAL PROTEIN | | Authors: | Cooray, S, Bahar, M.W, Abrescia, N.G.A, McVey, C.E, Bartlett, N.W, Chen, R.A.-J, Stuart, D.I, Grimes, J.M, Smith, G.L. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional and Structural Studies of the Vaccinia Virus Virulence Factor N1 Reveal a Bcl-2-Like Anti- Apoptotic Protein
J.Gen.Virol., 88, 2007
|
|
1UV6
 
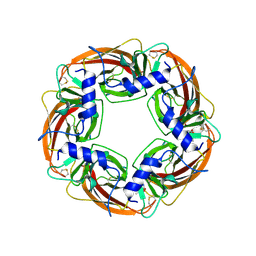 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with carbamylcholine | | Descriptor: | 2-[(AMINOCARBONYL)OXY]-N,N,N-TRIMETHYLETHANAMINIUM, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-01-15 | | Release date: | 2004-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
