8F93
 
 | | WDR5 covalently modified at Y228 by (R)-2-SF | | Descriptor: | 3-ethynyl-5-{[(3R)-4-{1-[(2-methoxyphenyl)methyl]-1H-benzimidazole-5-carbonyl}-3-methylpiperazin-1-yl]methyl}benzene-1-sulfonyl fluoride, CHLORIDE ION, GLYCEROL, ... | | Authors: | Taunton, J, Craven, G.B, Chen, Y. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Direct mapping of ligandable tyrosines and lysines in cells with chiral sulfonyl fluoride probes.
Nat.Chem., 15, 2023
|
|
4YHF
 
 | | Bruton's tyrosine kinase in complex with a t-butyl cyanoacrylamide inhibitor | | Descriptor: | (2S)-2-({(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}carbonyl)-4,4-dimethylpentanenitrile, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Paavilainen, V.O, McFarland, J.M, Taunton, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prolonged and tunable residence time using reversible covalent kinase inhibitors.
Nat.Chem.Biol., 11, 2015
|
|
9AUU
 
 | | Hsp90 NTD in complex with compound 6 | | Descriptor: | 3-{[(3R)-3-({6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}methyl)piperidin-1-yl]methyl}-6-fluoro-2-hydroxybenzaldehyde, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Weaver, J.M, Craven, G.B, Taunton, J. | | Deposit date: | 2024-02-29 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminomethyl Salicylaldehydes Lock onto a Surface Lysine by Forming an Extended Intramolecular Hydrogen Bond Network.
J.Am.Chem.Soc., 146, 2024
|
|
9C1W
 
 | | Structure of AKT2 with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 4-{2-[({4-[(2P)-2-(2-aminopyridin-3-yl)-5-phenyl-3H-imidazo[4,5-b]pyridin-3-yl]phenyl}methyl)amino]ethyl}-2-hydroxybenzaldehyde, RAC-beta serine/threonine-protein kinase | | Authors: | Craven, G.B, Ma, X, Taunton, J. | | Deposit date: | 2024-05-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutant-selective AKT inhibition through lysine targeting and neo-zinc chelation.
Nature, 637, 2025
|
|
4JG6
 
 | | RSK2 CTD bound to 2-cyano-3-(1H-indazol-5-yl)acrylamide | | Descriptor: | (2S)-2-cyano-3-(1H-indazol-5-yl)propanamide, Ribosomal protein S6 kinase alpha-3, SODIUM ION | | Authors: | Miller, R.M, Paavilainen, V.O, Krishnan, S, Serafimova, I.M, Taunton, J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-04-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Electrophilic fragment-based design of reversible covalent kinase inhibitors.
J.Am.Chem.Soc., 135, 2013
|
|
7FIC
 
 | |
8UW9
 
 | | Structure of AKT1(E17K) with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-({4-[(2P)-2-(2-aminopyridin-3-yl)-5-phenyl-3H-imidazo[4,5-b]pyridin-3-yl]phenyl}methyl)-2-(2-fluoro-4-formyl-3-hydroxyphenyl)acetamide, ... | | Authors: | Craven, G.B, Taunton, J. | | Deposit date: | 2023-11-06 | | Release date: | 2024-09-04 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutant-selective AKT inhibition through lysine targeting and neo-zinc chelation.
Nature, 637, 2025
|
|
8UW2
 
 | | Structure of AKT1(E17K) with compound 3 (zinc-free) | | Descriptor: | 1,2-ETHANEDIOL, 4-{2-[({4-[(2P)-2-(2-aminopyridin-3-yl)-5-phenyl-3H-imidazo[4,5-b]pyridin-3-yl]phenyl}methyl)amino]ethyl}-2-hydroxybenzaldehyde, NB41, ... | | Authors: | Craven, G.B, Taunton, J. | | Deposit date: | 2023-11-05 | | Release date: | 2024-09-04 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutant-selective AKT inhibition through lysine targeting and neo-zinc chelation.
Nature, 637, 2025
|
|
8UVY
 
 | | Structure of AKT1(E17K) with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 4-{2-[({4-[(2P)-2-(2-aminopyridin-3-yl)-5-phenyl-3H-imidazo[4,5-b]pyridin-3-yl]phenyl}methyl)amino]ethyl}-2-hydroxybenzaldehyde, NB41, ... | | Authors: | Craven, G.B, Taunton, J. | | Deposit date: | 2023-11-05 | | Release date: | 2024-09-04 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Mutant-selective AKT inhibition through lysine targeting and neo-zinc chelation.
Nature, 637, 2025
|
|
8UW7
 
 | | Structure of AKT1(WT) with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 4-{2-[({4-[(2P)-2-(2-aminopyridin-3-yl)-5-phenyl-3H-imidazo[4,5-b]pyridin-3-yl]phenyl}methyl)amino]ethyl}-2-hydroxybenzaldehyde, NB41, ... | | Authors: | Craven, G.B, Taunton, J. | | Deposit date: | 2023-11-06 | | Release date: | 2024-09-04 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Mutant-selective AKT inhibition through lysine targeting and neo-zinc chelation.
Nature, 637, 2025
|
|
5LZU
 
 | | Structure of the mammalian ribosomal termination complex with accommodated eRF1 | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5LZT
 
 | | Structure of the mammalian ribosomal termination complex with eRF1 and eRF3. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5LZS
 
 | | Structure of the mammalian ribosomal elongation complex with aminoacyl-tRNA, eEF1A, and didemnin B | | Descriptor: | (2~{S})-~{N}-[(2~{R})-1-[[(3~{S},6~{S},8~{S},12~{S},13~{R},16~{S},17~{R},20~{S},23~{S})-13-[(2~{S})-butan-2-yl]-20-[(4-methoxyphenyl)methyl]-6,17,21-trimethyl-3-(2-methylpropyl)-12-oxidanyl-2,5,7,10,15,19,22-heptakis(oxidanylidene)-8-propan-2-yl-9,18-dioxa-1,4,14,21-tetrazabicyclo[21.3.0]hexacosan-16-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-~{N}-methyl-1-[(2~{S})-2-oxidanylpropanoyl]pyrrolidine-2-carboxamide, 18S ribosomal RNA, 28S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
4D9T
 
 | | Rsk2 C-terminal Kinase Domain with inhibitor (E)-methyl 3-(4-amino-7-(3-hydroxypropyl)-5-p-tolyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-2-cyanoacrylate | | Descriptor: | Ribosomal protein S6 kinase alpha-3, SODIUM ION, methyl (2S)-3-{4-amino-7-[(1E)-3-hydroxyprop-1-en-1-yl]-5-(4-methylphenyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl}-2-cyanopropanoate | | Authors: | Serafimova, I.M, Pufall, M.A, Krishnan, S, Duda, K, Cohen, M.S, Maglathlin, R.L, McFarland, J.M, Miller, R.M, Frodin, M, Taunton, J. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles.
Nat.Chem.Biol., 8, 2012
|
|
4D9U
 
 | | Rsk2 C-terminal Kinase Domain, (E)-tert-butyl 3-(4-amino-7-(3-hydroxypropyl)-5-p-tolyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-2-cyanoacrylate | | Descriptor: | Ribosomal protein S6 kinase alpha-3, SODIUM ION, tert-butyl (2S)-3-[4-amino-7-(3-hydroxypropyl)-5-(4-methylphenyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl]-2-cyanopropanoate | | Authors: | Serafimova, I.M, Pufall, M.A, Krishnan, S, Duda, K, Cohen, M.S, Maglathlin, R.L, McFarland, J.M, Miller, R.M, Frodin, M, Taunton, J. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles.
Nat.Chem.Biol., 8, 2012
|
|
5LZY
 
 | | Structure of the mammalian rescue complex with Pelota and Hbs1l assembled on a polyadenylated mRNA. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5LZZ
 
 | | Structure of the mammalian rescue complex with Pelota and Hbs1l (combined) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5LZV
 
 | | Structure of the mammalian ribosomal termination complex with accommodated eRF1(AAQ) and ABCE1. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5LZX
 
 | | Structure of the mammalian rescue complex with Pelota and Hbs1l assembled on a UGA stop codon. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5LZW
 
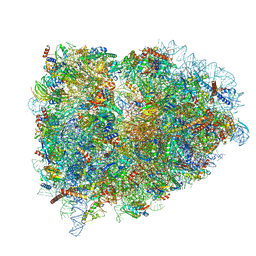 | | Structure of the mammalian rescue complex with Pelota and Hbs1l assembled on a truncated mRNA. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5K9I
 
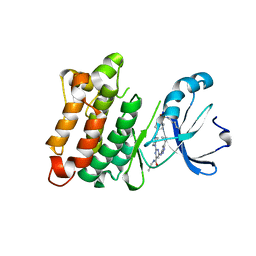 | | Crystal structure of c-SRC in complex with a covalent lysine probe | | Descriptor: | 4-[(4-{4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]-6-[(prop-2-yn-1-yl)carbamoyl]pyrimidin-2-yl}piperazin-1-yl)methyl]benzene-1-sulfonyl fluoride, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Wan, X, Ouyang, S, Zhao, Q, Taunton, J. | | Deposit date: | 2016-05-31 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes.
J. Am. Chem. Soc., 139, 2017
|
|
6U09
 
 | |
6U98
 
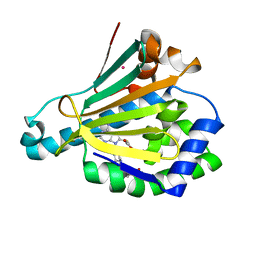 | | Hsp90a NTD K58R bound reversibly to sulfonyl fluoride 6 | | Descriptor: | 3-{[(3R)-3-({6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}methyl)piperidin-1-yl]methyl}benzene-1-sulfonyl fluoride, Heat shock protein HSP 90-alpha, POTASSIUM ION, ... | | Authors: | Cuesta, A, Wan, X, Taunton, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand Conformational Bias Drives Enantioselective Modification of a Surface-Exposed Lysine on Hsp90.
J.Am.Chem.Soc., 142, 2020
|
|
6U9B
 
 | | Hsp90a NTD covalently bound to sulfonyl fluoride 5 at K58 | | Descriptor: | 3-{[(3S)-3-({6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}methyl)piperidin-1-yl]methyl}benzene-1-sulfonyl fluoride, Heat shock protein HSP 90-alpha | | Authors: | Cuesta, A, Wan, X, Taunton, J. | | Deposit date: | 2019-09-07 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ligand Conformational Bias Drives Enantioselective Modification of a Surface-Exposed Lysine on Hsp90.
J.Am.Chem.Soc., 142, 2020
|
|
6U06
 
 | |
