7UI1
 
 | | Pfs230 D1D2 domain in complex with 230AL-37 | | Descriptor: | 230AL-37, Gametocyte surface protein P230, SULFATE ION | | Authors: | Tang, W.K, Tolia, N.H. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A human antibody epitope map of Pfs230D1 derived from analysis of individuals vaccinated with a malaria transmission-blocking vaccine.
Immunity, 56, 2023
|
|
7UCQ
 
 | | Pfs230 D1 domain in complex with 230AS-18 | | Descriptor: | 230AS-18, Gametocyte surface protein P230 | | Authors: | Tang, W.K, Tolia, N.H. | | Deposit date: | 2022-03-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A human antibody epitope map of Pfs230D1 derived from analysis of individuals vaccinated with a malaria transmission-blocking vaccine.
Immunity, 56, 2023
|
|
6XLY
 
 | |
1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
3TN2
 
 | | structure analysis of MIP1-beta P8A | | Descriptor: | C-C motif chemokine 4, ZINC ION | | Authors: | Guo, Q, Tang, W.J. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-05 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
2G54
 
 | |
2G47
 
 | |
2G48
 
 | |
2G49
 
 | |
8H9Z
 
 | | Annexin A5 protein mutant | | Descriptor: | Annexin A5, CALCIUM ION | | Authors: | Hua, Z.C, Tang, W. | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | structure dissection of the membrane aggregation mechanism induced by Annexin A5 mutation
To Be Published
|
|
7RZE
 
 | | Insulin Degrading Enzyme pO/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZI
 
 | | Insulin Degrading Enzyme pC/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZH
 
 | | Insulin Degrading Enzyme O/O | | Descriptor: | Cysteine-free Insulin-degrading enzyme | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZG
 
 | | Insulin Degrading Enzyme O/pO | | Descriptor: | Cysteine-free Insulin-degrading enzyme | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZF
 
 | | Insulin Degrading Enzyme O/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
4F33
 
 | | Crystal Structure of therapeutic antibody MORAb-009 | | Descriptor: | MORAb-009 FAB heavy chain, MORAb-009 FAB light chain, TETRAETHYLENE GLYCOL | | Authors: | Xia, D, Ma, J, Tang, W.K, Esser, L. | | Deposit date: | 2012-05-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Recognition of mesothelin by the therapeutic antibody MORAb-009: structural and mechanistic insights.
J.Biol.Chem., 287, 2012
|
|
6XOS
 
 | |
6XOU
 
 | |
6XOV
 
 | |
6XOT
 
 | |
6VHJ
 
 | |
6VGT
 
 | |
6VE9
 
 | |
6KY6
 
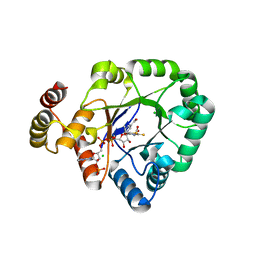 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complexs with inhibitor epalrestat in space group P3221cc | | Descriptor: | 2,5-diketo-D-gluconic acid reductase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, C.Y, Min, Z.Z, Liu, X.M, Wang, C, Tang, W.R. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
4F3F
 
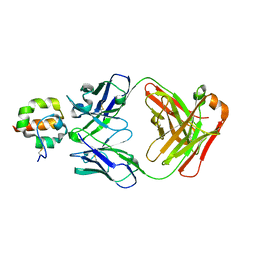 | | Crystal Structure of Msln7-64 MORAb-009 FAB complex | | Descriptor: | MORAb-009 Fab heavy chain, MORAb-009 Fab light chain, Mesothelin | | Authors: | Xia, D, Pastan, I, Ma, J, Tang, W.K, Esser, L. | | Deposit date: | 2012-05-09 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Recognition of mesothelin by the therapeutic antibody MORAb-009: structural and mechanistic insights.
J.Biol.Chem., 287, 2012
|
|
