2X6L
 
 | |
2X69
 
 | |
1XFW
 
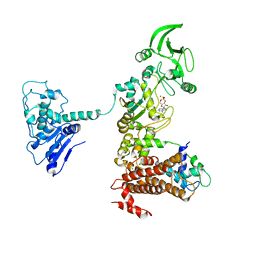 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and 3'5' cyclic AMP (cAMP) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CALCIUM ION, Calmodulin 2, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XFV
 
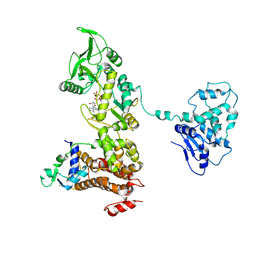 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and 3' deoxy-ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Calmodulin 2, ... | | Authors: | Shen, Q, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
2WK3
 
 | |
1XFX
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin in the presence of 10 millimolar exogenously added calcium chloride | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XFZ
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin in the presence of 1 millimolar exogenously added calcium chloride | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
4L3T
 
 | | Crystal Structure of Substrate-free Human Presequence Protease | | Descriptor: | ACETATE ION, GLYCEROL, Presequence protease, ... | | Authors: | King, J.V, Liang, W.G, Tang, W.J. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-03 | | Last modified: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis of substrate recognition and degradation by human presequence protease.
Structure, 22, 2014
|
|
2G47
 
 | |
2G48
 
 | |
3CWW
 
 | | Crystal Structure of IDE-bradykinin complex | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETATE ION, Insulin-degrading enzyme, ... | | Authors: | Malito, E, Tang, W.J. | | Deposit date: | 2008-04-23 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Molecular Bases for the Recognition of Short Peptide Substrates and Cysteine-Directed Modifications of Human Insulin-Degrading Enzyme
Biochemistry, 47, 2008
|
|
2G54
 
 | |
2G56
 
 | |
2G49
 
 | |
4RA8
 
 | | Structure analysis of the Mip1a P8A mutant | | Descriptor: | C-C motif chemokine 3 | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
4RAL
 
 | | Crystal structure of insulin degrading enzyme in complex with macrophage inflammatory protein 1 beta | | Descriptor: | C-C motif chemokine 4, Insulin-degrading enzyme, ZINC ION | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
1I5T
 
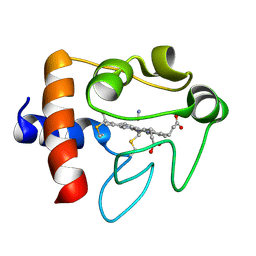 | | SOLUTION STRUCTURE OF CYANOFERRICYTOCHROME C | | Descriptor: | CYANIDE ION, CYTOCHROME C, HEME C | | Authors: | Yao, Y, Qian, C, Ye, K, Wang, J, Tang, W. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cyanoferricytochrome c: ligand-controlled conformational flexibility and electronic structure of the heme moiety.
J.Biol.Inorg.Chem., 7, 2002
|
|
4IOF
 
 | | Crystal structure analysis of Fab-bound human Insulin Degrading Enzyme (IDE) | | Descriptor: | Fab-bound IDE, heavy chain, light chain, ... | | Authors: | McCord, L.A, Liang, W.G, Hoey, R, Dowdell, E, Koide, A, Koide, S, Tang, W.J. | | Deposit date: | 2013-01-07 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.353 Å) | | Cite: | Conformational states and recognition of amyloidogenic peptides of human insulin-degrading enzyme.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1K93
 
 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, CALMODULIN-SENSITIVE ADENYLATE CYCLASE, ... | | Authors: | Drum, C.L, Yan, S.-Z, Bard, J, Shen, Y.-Q, Lu, D, Soelaiman, S, Grabarek, Z, Bohm, A, Tang, W.-J. | | Deposit date: | 2001-10-26 | | Release date: | 2002-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin.
Nature, 415, 2002
|
|
8TWF
 
 | |
1M60
 
 | | Solution Structure of Zinc-substituted cytochrome c | | Descriptor: | ZINC SUBSTITUTED HEME C, Zinc-substituted cytochrome c | | Authors: | Qian, C, Yao, Y, Tong, Y, Wang, J, Tang, W. | | Deposit date: | 2002-07-11 | | Release date: | 2002-08-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of zinc-substituted cytochrome c.
J.Biol.Inorg.Chem., 8, 2003
|
|
8J23
 
 | | Cryo-EM structure of FFAR2 complex in apo state | | Descriptor: | Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short chain fatty acid receptors FFAR2 and FFAR3
To Be Published
|
|
4QIA
 
 | | Crystal structure of human insulin degrading enzyme (ide) in complex with inhibitor N-benzyl-N-(carboxymethyl)glycyl-L-histidine | | Descriptor: | Insulin-degrading enzyme, N-benzyl-N-(carboxymethyl)glycyl-L-histidine, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structure-activity relationships of imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, dual binders of human insulin-degrading enzyme.
Eur.J.Med.Chem., 90, 2015
|
|
5CJO
 
 | |
7EHW
 
 | | BRD4-BD1 in complex with LT-642-602 | | Descriptor: | 1-[4-ethyl-2-methyl-5-(6-morpholin-4-yl-1H-benzimidazol-2-yl)-1H-pyrrol-3-yl]ethanone, Bromodomain-containing protein 4 | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | BRD4-BD1 in complex with LT-642-602
To Be Published
|
|
