5KLV
 
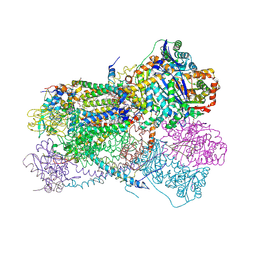 | | Structure of bos taurus cytochrome bc1 with fenamidone inhibited | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, (5S)-5-methyl-2-(methylsulfanyl)-5-phenyl-3-(phenylamino)-3,5-dihydro-4H-imidazol-4-one, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Zhou, Y, Xiao, Y, Tang, W.K, Yu, C.A, Qin, Z. | | Deposit date: | 2016-06-25 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
6IXN
 
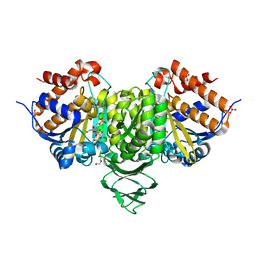 | | Crystal structure of isocitrate dehydrogenase from Ostreococcus tauri in complex with NAD+ and citrate | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhu, G.P, Tang, W.G, Wang, P. | | Deposit date: | 2018-12-11 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of NAD + -linked isocitrate dehydrogenase from the green alga Ostreococcus tauri and its evolutionary relationship with eukaryotic NADP + -linked homologs.
Arch.Biochem.Biophys., 708, 2021
|
|
7JUM
 
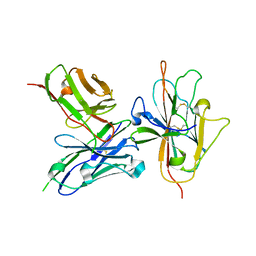 | | Pfs230 D1 domain in complex with neutralizing antibody LMIV230-01 | | Descriptor: | Gametocyte surface protein P230, Neutralizing antibody, LMIV230-01 Single-chain Fv | | Authors: | Tolia, N, Tang, W.K. | | Deposit date: | 2020-08-20 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | A human monoclonal antibody blocks malaria transmission and defines a highly conserved neutralizing epitope on gametes.
Nat Commun, 12, 2021
|
|
6IXT
 
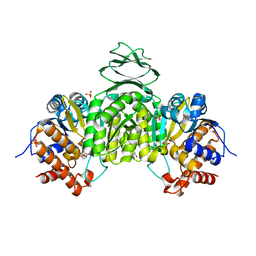 | | Crystal structure of isocitrate dehydrogenase from Ostreococcus tauri in complex with NAD+ and Mg2+ | | Descriptor: | GLYCEROL, Isocitrate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Zhu, G.P, Tang, W.G, Wang, P. | | Deposit date: | 2018-12-12 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of NAD + -linked isocitrate dehydrogenase from the green alga Ostreococcus tauri and its evolutionary relationship with eukaryotic NADP + -linked homologs.
Arch.Biochem.Biophys., 708, 2021
|
|
7K1D
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_77291 | | Descriptor: | (3R)-3-{4-[(3R)-4-(3,4-difluorobenzene-1-carbonyl)morpholin-3-yl]-1H-1,2,3-triazol-1-yl}-N-hydroxy-4-(naphthalen-2-yl)butanamide, 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 2
To Be Published
|
|
7K1F
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_88558 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 3,4-difluoro-N-({1-[(2R)-4-(hydroxyamino)-4-oxo-1-(quinolin-7-yl)butan-2-yl]-1H-1,2,3-triazol-4-yl}methyl)benzamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 4
To Be Published
|
|
7K1E
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_88646 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 3,4-difluoro-N-[(1S)-1-{1-[(2R)-4-(hydroxyamino)-4-oxo-1-(5,6,7,8-tetrahydronaphthalen-2-yl)butan-2-yl]-1H-1,2,3-triazol-4-yl}ethyl]benzamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 3
To Be Published
|
|
4MHE
 
 | | Crystal structure of CC-chemokine 18 | | Descriptor: | ACETATE ION, C-C motif chemokine 18 | | Authors: | Liang, W.G, Tang, W.-J. | | Deposit date: | 2013-08-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
3E4Z
 
 | | Crystal structure of human insulin degrading enzyme in complex with insulin-like growth factor II | | Descriptor: | Insulin-degrading enzyme, Insulin-like growth factor II, ZINC ION | | Authors: | Guo, Q, Manolopoulou, M, Tang, W.-J. | | Deposit date: | 2008-08-12 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular Basis for the Recognition and Cleavages of IGF-II, TGF-alpha, and Amylin by Human Insulin-Degrading Enzyme.
J.Mol.Biol., 395, 2010
|
|
4NXO
 
 | | Crystal Structure of Insulin Degrading Enzyme in complex with BDM44768 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Deprez, B, Tang, W. | | Deposit date: | 2013-12-09 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
6IXL
 
 | | Crystal structure of isocitrate dehydrogenase from Ostreococcus tauri | | Descriptor: | GLYCEROL, Isocitrate dehydrogenase, SULFATE ION | | Authors: | Zhu, G.P, Tang, W.G, Wang, P. | | Deposit date: | 2018-12-11 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of NAD + -linked isocitrate dehydrogenase from the green alga Ostreococcus tauri and its evolutionary relationship with eukaryotic NADP + -linked homologs.
Arch.Biochem.Biophys., 708, 2021
|
|
3E50
 
 | | Crystal structure of human insulin degrading enzyme in complex with transforming growth factor-alpha | | Descriptor: | Insulin-degrading enzyme, Protransforming growth factor alpha, ZINC ION | | Authors: | Guo, Q, Manolopoulou, M, Tang, W.-J. | | Deposit date: | 2008-08-12 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for the Recognition and Cleavages of IGF-II, TGF-alpha, and Amylin by Human Insulin-Degrading Enzyme.
J.Mol.Biol., 395, 2010
|
|
1S26
 
 | | Structure of Anthrax Edema Factor-Calmodulin-alpha,beta-methyleneadenosine 5'-triphosphate Complex Reveals an Alternative Mode of ATP Binding to the Catalytic Site | | Descriptor: | CALCIUM ION, Calmodulin, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Bohm, A, Tang, W.-J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of anthrax edema factor-calmodulin-adenosine-5'-(alpha,beta-methylene)-triphosphate complex reveals an alternative mode of ATP binding to the catalytic site
Biochem.Biophys.Res.Commun., 317, 2004
|
|
8JPP
 
 | | Cryo-EM structure of succinate receptor bound to succinate acid coupling MiniGsq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Wang, T.X, Tang, W.Q, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-06-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular activation and G protein coupling selectivity of human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
8JPN
 
 | | Cryo-EM structure of succinate receptor bound to cis-epoxysuccinic acid coupling to Gi | | Descriptor: | (2R,3S)-oxirane-2,3-dicarboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, T.X, Tang, W.Q, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-06-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular activation and G protein coupling selectivity of human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
5CJO
 
 | |
1K90
 
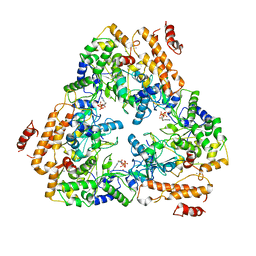 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin and 3' deoxy-ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CALMODULIN, ... | | Authors: | Drum, C.L, Yan, S.-Z, Bard, J, Shen, Y.-Q, Lu, D, Soelaiman, S, Grabarek, Z, Bohm, A, Tang, W.-J. | | Deposit date: | 2001-10-26 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin.
Nature, 415, 2002
|
|
5C6R
 
 | | Crystal structure of PH domain of ASAP1 | | Descriptor: | Arf-GAP, PHOSPHATE ION, TRIETHYLENE GLYCOL | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2015-06-23 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Cooperative Binding of Anionic Phospholipids to the PH Domain of the Arf GAP ASAP1.
Structure, 23, 2015
|
|
4IOF
 
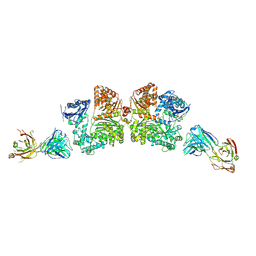 | | Crystal structure analysis of Fab-bound human Insulin Degrading Enzyme (IDE) | | Descriptor: | Fab-bound IDE, heavy chain, light chain, ... | | Authors: | McCord, L.A, Liang, W.G, Hoey, R, Dowdell, E, Koide, A, Koide, S, Tang, W.J. | | Deposit date: | 2013-01-07 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.353 Å) | | Cite: | Conformational states and recognition of amyloidogenic peptides of human insulin-degrading enzyme.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8TWF
 
 | |
1NMJ
 
 | |
4GSF
 
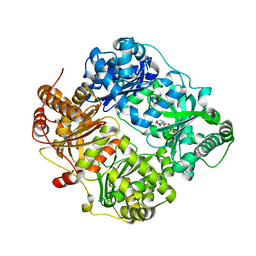 | | The structure analysis of cysteine free insulin degrading enzyme (ide) with (s)-2-{2-[carboxymethyl-(3-phenyl-propionyl)-amino]-acetylamino}-3-(3h-imidazol-4-yl)-propionic acid methyl ester | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl N-(carboxymethyl)-N-(3-phenylpropanoyl)glycyl-D-histidinate | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-activity relationships of imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, dual binders of human insulin-degrading enzyme.
Eur.J.Med.Chem., 90, 2015
|
|
5C79
 
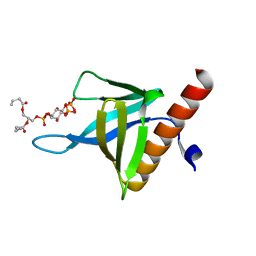 | | PH domain of ASAP1 in complex with diC4-PtdIns(4,5)P2 | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, Arf-GAP, CHLORIDE ION | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2015-06-24 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Basis for Cooperative Binding of Anionic Phospholipids to the PH Domain of the Arf GAP ASAP1.
Structure, 23, 2015
|
|
1XFZ
 
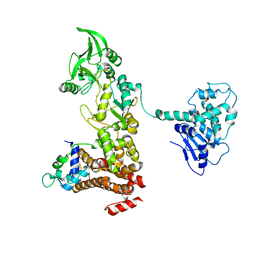 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin in the presence of 1 millimolar exogenously added calcium chloride | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
6B70
 
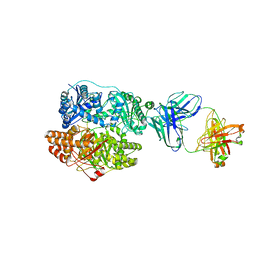 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain and insulin | | Descriptor: | FAB H11-E heavy chain, FAB H11-E light chain, Insulin, ... | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
