5KMK
 
 | |
5KMN
 
 | |
5KZ8
 
 | | Mark2 complex with 7-[(1S)-1-(4-fluorophenyl)ethyl]-5,5-dimethyl-2-(3-pyridylamino)pyrrolo[2,3-d]pyrimidin-6-one | | Descriptor: | 5,5-dimethyl-7-[(1~{S})-4-oxidanyl-1~{H}-inden-1-yl]-2-phenylazanyl-pyrrolo[2,3-d]pyrimidin-6-one, Serine/threonine-protein kinase MARK2 | | Authors: | Su, H.P, Munshi, S.K. | | Deposit date: | 2016-07-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structure guided design of a series of selective pyrrolopyrimidinone MARK inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5KMO
 
 | | TrkA JM-kinase with 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-(2-pyridyl)urea | | Descriptor: | 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-pyridin-2-yl-urea, High affinity nerve growth factor receptor | | Authors: | Su, H.P. | | Deposit date: | 2016-06-27 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural characterization of nonactive site, TrkA-selective kinase inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KMM
 
 | | TrkA JM-kinase with 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-(1-naphthyl)urea | | Descriptor: | 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-naphthalen-1-yl-urea, High affinity nerve growth factor receptor | | Authors: | Su, H.P. | | Deposit date: | 2016-06-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural characterization of nonactive site, TrkA-selective kinase inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KMI
 
 | |
5KZ7
 
 | | Mark2 complex with 7-[(1S)-1-(4-fluorophenyl)ethyl]-5,5-dimethyl-2-(3-pyridylamino)pyrrolo[2,3-d]pyrimidin-6-one | | Descriptor: | 7-[(1~{S})-1-(4-fluorophenyl)ethyl]-5,5-dimethyl-2-(pyridin-3-ylamino)pyrrolo[2,3-d]pyrimidin-6-one, Serine/threonine-protein kinase MARK2 | | Authors: | Su, H.P, Munshi, S.K. | | Deposit date: | 2016-07-23 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure guided design of a series of selective pyrrolopyrimidinone MARK inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5KMJ
 
 | | TrkA JM-kinase with {N}-(2-pyridylmethyl)-2-[2-(2-thienyl)indol-1-yl]acetamide | | Descriptor: | High affinity nerve growth factor receptor, ~{N}-(pyridin-2-ylmethyl)-2-(2-thiophen-2-ylindol-1-yl)ethanamide | | Authors: | Su, H.P. | | Deposit date: | 2016-06-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural characterization of nonactive site, TrkA-selective kinase inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5IVQ
 
 | |
2I9L
 
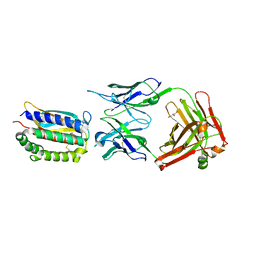 | | Structure of Fab 7D11 from a neutralizing antibody against the poxvirus L1 protein | | Descriptor: | Antibody 7D11 heavy chain, Antibody 7D11 light chain, GLYCEROL, ... | | Authors: | Su, H.P, Golden, J.W, Gittis, A.G, Moss, B, Hooper, J.W, Garboczi, D.N. | | Deposit date: | 2006-09-05 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the binding of the neutralizing antibody, 7D11, to the poxvirus L1 protein
Virology, 368, 2007
|
|
5HX8
 
 | |
5I8A
 
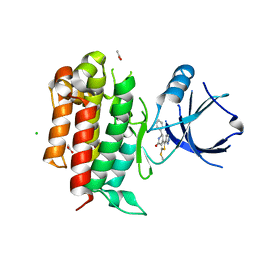 | | TrkA with (6~{R})-3-methylsulfanyl-6-phenyl-1-(1~{H}-pyrazol-3-yl)-6,7-dihydro-5~{H}-thieno[3,4-c]pyridin-4-one | | Descriptor: | (6R)-3-(methylsulfanyl)-6-phenyl-1-(1H-pyrazol-3-yl)-6,7-dihydrothieno[3,4-c]pyridin-4(5H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Su, H.P. | | Deposit date: | 2016-02-18 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Potent, selective and orally bioavailable leucine-rich repeat kinase 2 (LRRK2) inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
3C64
 
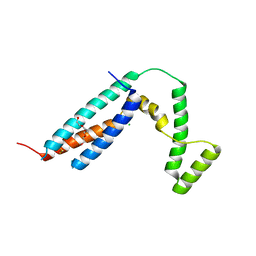 | | The MC179 portion of the Cysteine-rich Interdomain Region (CIDR) of a Plasmodium falciparum Erythrocyte Membrane Protein-1 (PfEMP1) | | Descriptor: | CHLORIDE ION, PfEMP1 variant 2 of strain MC, TETRAETHYLENE GLYCOL | | Authors: | Klein, M.M, Gittis, A.G, Su, H.P, Makobongo, M.O, Moore, J.M, Singh, S, Miller, L.H, Garboczi, D.N. | | Deposit date: | 2008-02-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Cysteine-Rich Interdomain Region from the Highly Variable Plasmodium falciparum Erythrocyte Membrane Protein-1 Exhibits a Conserved Structure.
Plos Pathog., 4, 2008
|
|
5VZ6
 
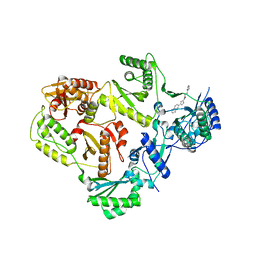 | | HIV Reverse Transcriptase complexed with (E)-3-(pyrimidin-2-yl)-N-(5-(5,6,7,8-tetrahydronaphthalen-2-yl)-1H-pyrazol-3-yl)acrylamide | | Descriptor: | 3-(pyrimidin-2-yl)-N-[3-(5,6,7,8-tetrahydronaphthalen-2-yl)-1H-pyrazol-5-yl]propanamide, HIV Reverse Transcriptase | | Authors: | Yan, Y, Su, H.P. | | Deposit date: | 2017-05-26 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | HIV Reverse Transcriptase complexed with inhibitor
To Be Published
|
|
1Z27
 
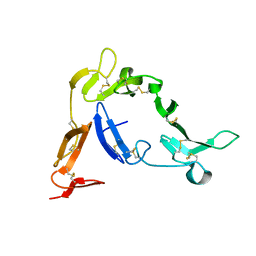 | | Crystal structure of Native Pvs25, an ookinete protein from Plasmodium vivax. | | Descriptor: | ookinete surface protein Pvs25 | | Authors: | Saxena, A.K, Singh, K, Su, H.P, Klein, M.M, Stowers, A.W, Saul, A.J, Long, C.A, Garboczi, D.N. | | Deposit date: | 2005-03-07 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The essential mosquito-stage P25 and P28 proteins from Plasmodium form tile-like triangular prisms
Nat.Struct.Mol.Biol., 13, 2006
|
|
1Z1Y
 
 | | Crystal structure of Methylated Pvs25, an ookinete protein from Plasmodium vivax | | Descriptor: | YTTERBIUM (III) ION, ookinete surface protein Pvs25 | | Authors: | Saxena, A.K, Singh, K, Su, H.P, Klein, M.M, Stowers, A.W, Saul, A.J, Long, C.A, Garboczi, D.N. | | Deposit date: | 2005-03-07 | | Release date: | 2005-12-06 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The essential mosquito-stage P25 and P28 proteins from Plasmodium form tile-like triangular prisms
Nat.Struct.Mol.Biol., 13, 2006
|
|
1Z3G
 
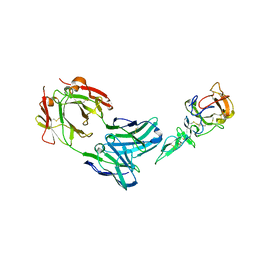 | | Crystal structure of complex between Pvs25 and Fab fragment of malaria transmission blocking antibody 2A8 | | Descriptor: | 2A8 Fab Heavy Chain, 2A8 Fab Light Chain, ookinete surface protein Pvs25 | | Authors: | Saxena, A.K, Singh, K, Su, H.P, Klein, M.M, Stowers, A.W, Saul, A.J, Long, C.A, Garboczi, D.N. | | Deposit date: | 2005-03-12 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The essential mosquito-stage P25 and P28 proteins from Plasmodium form tile-like triangular prisms
Nat.Struct.Mol.Biol., 13, 2006
|
|
3BIK
 
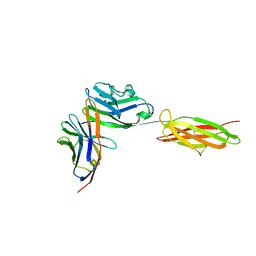 | | Crystal Structure of the PD-1/PD-L1 Complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BIS
 
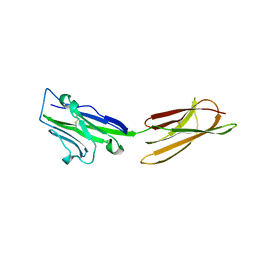 | | Crystal Structure of the PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5EJ0
 
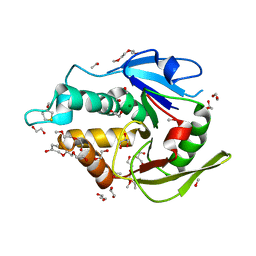 | | The vaccinia virus H3 envelope protein, a major target of neutralizing antibodies, exhibits a glycosyltransferase fold and binds UDP-Glucose | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Singh, K, Gittis, A.G, Gitti, R.K, Ostazesky, S.A, Su, H.P, Garboczi, D.N. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Vaccinia Virus H3 Envelope Protein, a Major Target of Neutralizing Antibodies, Exhibits a Glycosyltransferase Fold and Binds UDP-Glucose.
J.Virol., 90, 2016
|
|
3LP3
 
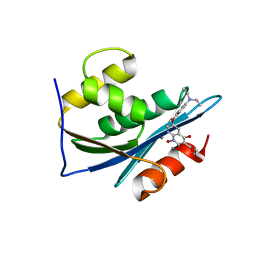 | | p15 HIV RNaseH domain with inhibitor MK3 | | Descriptor: | 3-[4-(diethylamino)phenoxy]-6-(ethoxycarbonyl)-5,8-dihydroxy-7-oxo-7,8-dihydro-1,8-naphthyridin-1-ium, MANGANESE (II) ION, p15 | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
3LP2
 
 | | HIV-1 reverse transcriptase with inhibitor | | Descriptor: | 3-[4-(diethylamino)phenoxy]-6-(ethoxycarbonyl)-5,8-dihydroxy-7-oxo-7,8-dihydro-1,8-naphthyridin-1-ium, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
3LP1
 
 | | HIV-1 reverse transcriptase with inhibitor | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 3-cyclopentyl-1,4-dihydroxy-1,8-naphthyridin-2(1H)-one, MANGANESE (II) ION, ... | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
3LP0
 
 | | HIV-1 reverse transcriptase with inhibitor | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
3RVG
 
 | | Crystals structure of Jak2 with a 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitor | | Descriptor: | 1-(cyclohexylamino)-7-(1-methyl-1H-pyrazol-4-yl)-5H-pyrido[4,3-b]indole-4-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Lim, J, Taoka, B, Otte, R.D, Spencer, K, Dinsmore, C.J, Altman, M.D, Chan, G, Rosenstein, C, Sharma, S, Su, H.P, Szewczak, A.A, Xu, L, Yin, H, Zugay-Murphy, J, Marshall, C.G, Young, J.R. | | Deposit date: | 2011-05-06 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Discovery of 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitors of Janus kinase 2 (JAK2) for the treatment of myeloproliferative disorders.
J.Med.Chem., 54, 2011
|
|
