5UXC
 
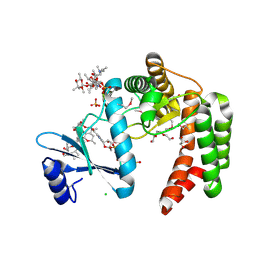 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with GDP | | Descriptor: | AZITHROMYCIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5UXD
 
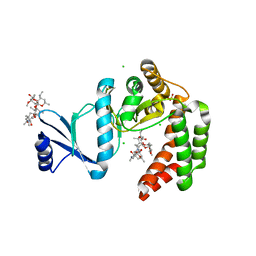 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with azithromycin | | Descriptor: | AZITHROMYCIN, CHLORIDE ION, Macrolide 2'-phosphotransferase MphH, ... | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5UXB
 
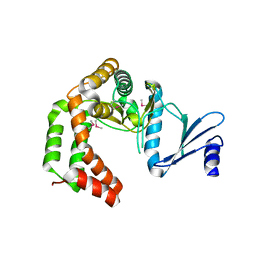 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium, apoenzyme | | Descriptor: | CHLORIDE ION, Macrolide 2'-phosphotransferase MphH | | Authors: | Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5US1
 
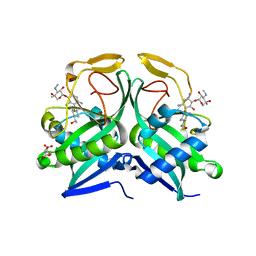 | | Crystal structure of aminoglycoside acetyltransferase AAC(2')-Ia in complex with N2'-acetylgentamicin C1A and coenzyme A | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-(acetylamino)-6-amino-2,3,4,6-tetradeoxy-alpha-D-erythro-hexopyranoside, ACETYL COENZYME *A, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Xu, Z, Wawrzak, Z, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Plazomicin Retains Antibiotic Activity against Most Aminoglycoside Modifying Enzymes.
ACS Infect Dis, 4, 2018
|
|
6OZ1
 
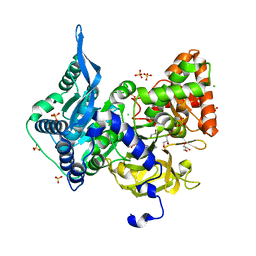 | | Crystal structure of the adenylation (A) domain of the carboxylate reductase (CAR) GR01_22995 from Mycobacterium chelonae | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Fedorchuk, T, Khusnutdinova, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | One-Pot Biocatalytic Transformation of Adipic Acid to 6-Aminocaproic Acid and 1,6-Hexamethylenediamine Using Carboxylic Acid Reductases and Transaminases.
J.Am.Chem.Soc., 142, 2020
|
|
7TOK
 
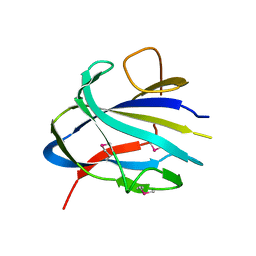 | | Crystal structure of the CBM domain of carbohydrate esterase FjoAcXE | | Descriptor: | Acetylxylan esterase I | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOG
 
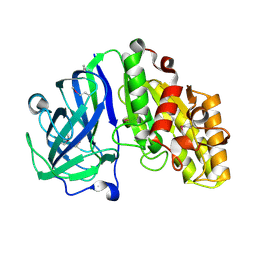 | | Crystal structure of carbohydrate esterase PbeAcXE, apoenzyme | | Descriptor: | SGNH hydrolase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOI
 
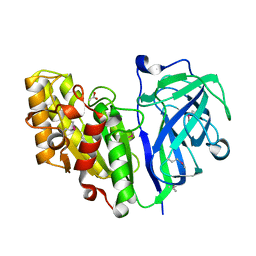 | | Crystal structure of carbohydrate esterase PbeAcXE, in complex with acetate | | Descriptor: | ACETATE ION, SGNH hydrolase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOJ
 
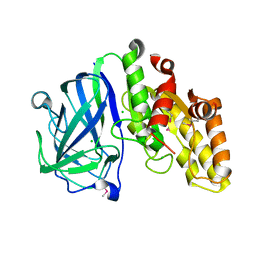 | | Crystal structure of carbohydrate esterase CspAcXE, apoenzyme | | Descriptor: | CHLORIDE ION, SGNH/GDSL hydrolase family protein | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOH
 
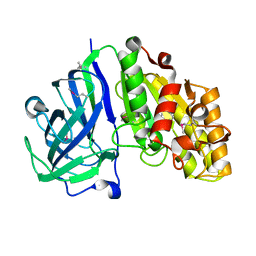 | | Crystal structure of carbohydrate esterase PbeAcXE, in complex with MeGlcpA-Xylp | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose, SGNH hydrolase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
4EBK
 
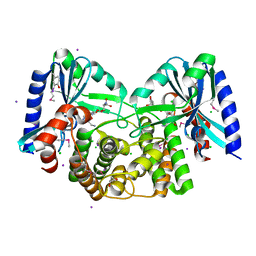 | | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, tobramycin-bound | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside nucleotidyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Dong, A, Minasov, G, Evdokimova, E, Egorova, O, Yim, V, Kudritska, M, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-23 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, tobramycin-bound
To be Published
|
|
4EJ7
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, ATP-bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aminoglycoside 3'-phosphotransferase AphA1-IAB, CALCIUM ION, ... | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Evdokimova, E, Egorova, O, Di Leo, R, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-18 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4E8O
 
 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-Ih from Acinetobacter baumannii | | Descriptor: | Aac(6')-Ih protein, CHLORIDE ION | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
4ECL
 
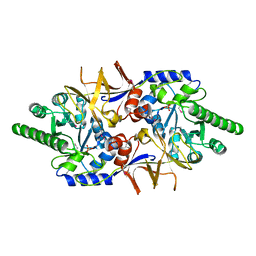 | | Crystal structure of the cytoplasmic domain of vancomycin resistance serine racemase VanTg | | Descriptor: | CHLORIDE ION, SULFATE ION, Serine racemase | | Authors: | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Cosme, J, Di Leo, R, Krishnamoorthy, M, Meziane-Cherif, D, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | Structural and Functional Adaptation of Vancomycin Resistance VanT Serine Racemases.
MBio, 6, 2015
|
|
4EBJ
 
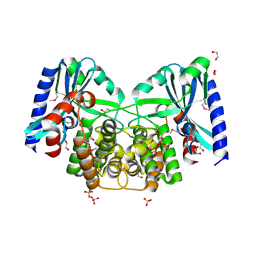 | | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, apo | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside nucleotidyltransferase, SULFATE ION | | Authors: | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Yim, V, Kudritska, M, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-23 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, apo
To be Published
|
|
4DE4
 
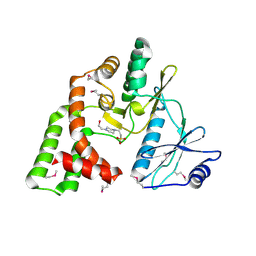 | | Crystal structure of aminoglycoside phosphotransferase APH(2")-Id/APH(2")-IVa in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, APH(2")-Id | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Nocek, B, Evdokimova, E, Egorova, O, Di Leo, R, Li, H, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
4DCA
 
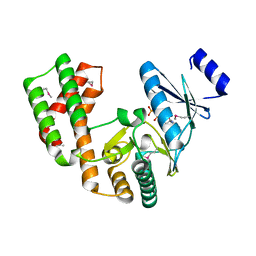 | | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, ADP-bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside phosphotransferase, MAGNESIUM ION | | Authors: | Stogios, P.J, Minasov, G, Singer, A.U, Tan, K, Nocek, B, Evdokimova, E, Egorova, O, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, ADP-bound
TO BE PUBLISHED
|
|
4EVY
 
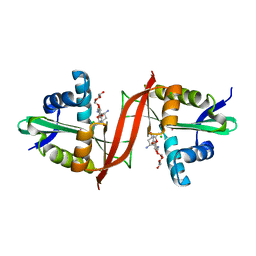 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-Ig from Acinetobacter haemolyticus in complex with tobramycin | | Descriptor: | Aminoglycoside N(6')-acetyltransferase type 1, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Minasov, G, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
4DFU
 
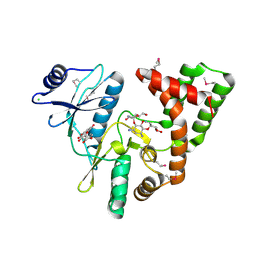 | | Inhibition of an antibiotic resistance enzyme: crystal structure of aminoglycoside phosphotransferase APH(2")-ID/APH(2")-IVA in complex with kanamycin inhibited with quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, APH(2")-Id, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Egorova, E, Di Leo, R, Li, H, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-24 | | Release date: | 2012-02-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
4DBX
 
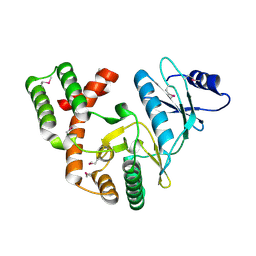 | | Crystal structure of aminoglycoside phosphotransferase APH(2")-ID/APH(2")-IVA | | Descriptor: | APH(2")-ID | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Nocek, B, Singer, A.U, Evdokimova, E, Egorova, E, Di Leo, R, Li, H, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-16 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
4F0Y
 
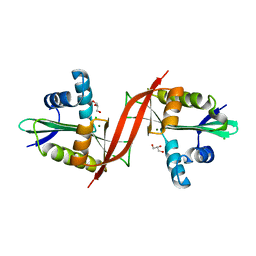 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-IG from Acinetobacter haemolyticus, apo | | Descriptor: | Aminoglycoside N(6')-acetyltransferase type 1, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Dong, A, Minasov, G, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-05 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
4DFB
 
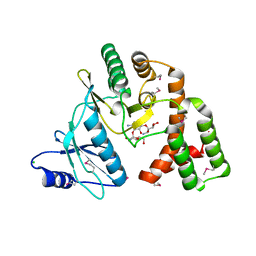 | | Crystal structure of aminoglycoside phosphotransferase aph(2")-id/aph(2")-iva in complex with kanamycin | | Descriptor: | APH(2")-Id, CHLORIDE ION, KANAMYCIN A | | Authors: | Stogios, P.J, Minasov, G, Osipiuk, J, Evdokimova, E, Egorova, E, Di leo, R, Li, H, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
4EXL
 
 | | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Phosphate-binding protein pstS 1 | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A
To be Published
|
|
4FEV
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor pyrazolopyrimidine PP1 | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4F78
 
 | | Crystal Structure of Vancomycin Resistance D,D-dipeptidase VanXYg | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Evdokimova, E, Minasov, G, Egorova, O, Di Leo, R, Kudritska, M, Yim, V, Meziane-Cherif, D, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
