3K84
 
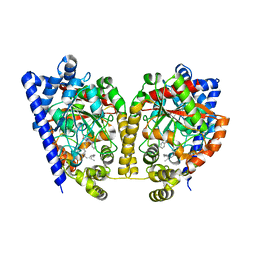 | | Crystal Structure Analysis of a Oleyl/Oxadiazole/pyridine Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | (9Z)-1-(5-pyridin-2-yl-1,3,4-oxadiazol-2-yl)octadec-9-en-1-one, CHLORIDE ION, Fatty-acid amide hydrolase 1 | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2009-10-13 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
3K83
 
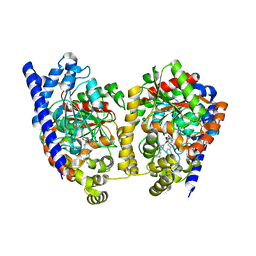 | | Crystal Structure Analysis of a Biphenyl/Oxazole/Carboxypyridine alpha-ketoheterocycle Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | 1-DODECANOL, 6-[2-(3-biphenyl-4-ylpropanoyl)-1,3-oxazol-5-yl]pyridine-2-carboxylic acid, CHLORIDE ION, ... | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2009-10-13 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
3K3Q
 
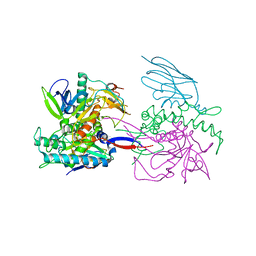 | | Crystal Structure of a Llama Antibody complexed with the C. Botulinum Neurotoxin Serotype A Catalytic Domain | | Descriptor: | Botulinum neurotoxin type A, ZINC ION, llama Aa1 VHH domain | | Authors: | Thompson, A.A, Dong, J, Marks, J.D, Stevens, R.C. | | Deposit date: | 2009-10-04 | | Release date: | 2010-02-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Single-Domain Llama Antibody Potently Inhibits the Enzymatic Activity of Botulinum Neurotoxin by Binding to the Non-Catalytic alpha-Exosite Binding Region.
J.Mol.Biol., 397, 2010
|
|
3K7F
 
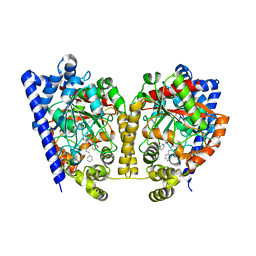 | | Crystal Structure Analysis of a Phenhexyl/Oxazole/Carboxypyridine alpha-Ketoheterocycle Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase' | | Descriptor: | 6-[2-(7-phenylheptanoyl)-1,3-oxazol-5-yl]pyridine-2-carboxylic acid, CHLORIDE ION, Fatty-acid amide hydrolase 1, ... | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2009-10-13 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
1HKL
 
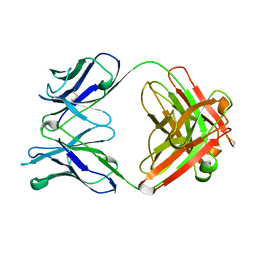 | | FREE AND LIGANDED FORM OF AN ESTEROLYTIC CATALYTIC ANTIBODY | | Descriptor: | 48G7 FAB | | Authors: | Wedemayer, G.J, Wang, L.H, Patten, P.A, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1996-12-20 | | Release date: | 1997-03-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structures of the free and liganded form of an esterolytic catalytic antibody.
J.Mol.Biol., 268, 1997
|
|
3LJ6
 
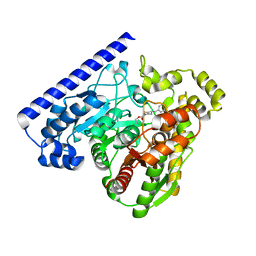 | |
3LJ7
 
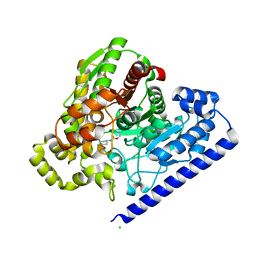 | |
3EML
 
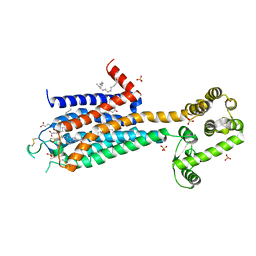 | | The 2.6 A Crystal Structure of a Human A2A Adenosine Receptor bound to ZM241385. | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Human Adenosine A2A receptor/T4 lysozyme chimera, STEARIC ACID, ... | | Authors: | Jaakola, V.-P, Griffith, M.T, Hanson, M.A, Cherezov, V, Chien, E.Y.T, Lane, J.R, Ijzerman, A.P, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2008-09-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist.
Science, 322, 2008
|
|
3FCT
 
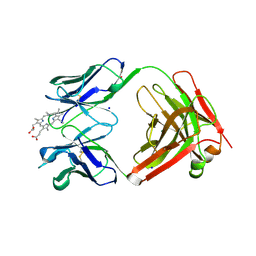 | | MATURE METAL CHELATASE CATALYTIC ANTIBODY WITH HAPTEN | | Descriptor: | CADMIUM ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Romesberg, F.E, Santarsiero, B.D, Barnes, D, Yin, J, Spiller, B, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and kinetic evidence for strain in biological catalysis.
Biochemistry, 37, 1998
|
|
3E1U
 
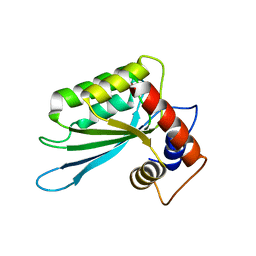 | | The Crystal Structure of the Anti-Viral APOBEC3G Catalytic Domain | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Holden, L, Prochnow, C, Chang, Y.P, Bransteitter, R, Chelico, L, Sen, U, Stevens, R.C, Goodman, R.F, Chen, X.S. | | Deposit date: | 2008-08-04 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications.
Nature, 456, 2008
|
|
4IAR
 
 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with ergotamine (PSI Community Target) | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Ergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
4J6S
 
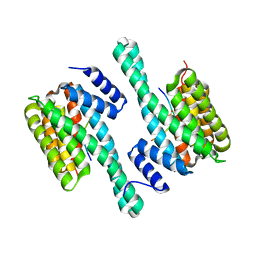 | |
4J5P
 
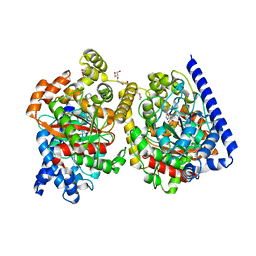 | | Crystal Structure of a Covalently Bound alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | (1S)-1-{5-[5-(bromomethyl)pyridin-2-yl]-1,3-oxazol-2-yl}-7-phenylheptan-1-ol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Otrubova, K, Brown, M, McCormick, M.S, Han, G.W, O'Neal, S.T, Cravatt, B.F, Stevens, R.C, Lichtman, A.H, Boger, D.L. | | Deposit date: | 2013-02-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design of Fatty Acid amide hydrolase inhibitors that act by covalently bonding to two active site residues.
J.Am.Chem.Soc., 135, 2013
|
|
4H37
 
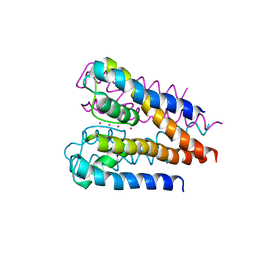 | | Crystal structure of a voltage-gated K+ channel pore domain in a closed state in lipid membranes | | Descriptor: | Lmo2059 protein, POTASSIUM ION | | Authors: | Santos, J.S, Asmar-Rovira, G.A, Han, G.W, Liu, W, Syeda, R, Cherezov, V, Baker, K.A, Stevens, R.C, Montal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of a Voltage-gated K+ Channel Pore Module in a Closed State in Lipid Membranes.
J.Biol.Chem., 287, 2012
|
|
4JKV
 
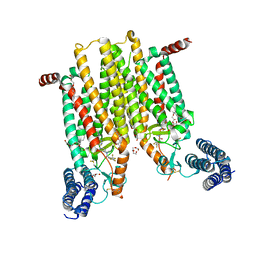 | | Structure of the human smoothened 7TM receptor in complex with an antitumor agent | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-fluoro-N-methyl-N-{1-[4-(1-methyl-1H-pyrazol-5-yl)phthalazin-1-yl]piperidin-4-yl}-2-(trifluoromethyl)benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, C, Wu, H, Katritch, V, Han, G.W, Huang, X, Liu, W, Siu, F.Y, Roth, B.L, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-03-11 | | Release date: | 2013-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the human smoothened receptor bound to an antitumour agent.
Nature, 497, 2013
|
|
4IAQ
 
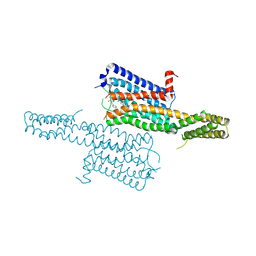 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with dihydroergotamine (PSI Community Target) | | Descriptor: | Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Dihydroergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
4IB4
 
 | | Crystal structure of the chimeric protein of 5-HT2B-BRIL in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, ... | | Authors: | Wacker, D, Wang, C, Katritch, V, Han, G.W, Huang, X, Vardy, E, McCorvy, J.D, Jiang, Y, Chu, M, Siu, F.Y, Liu, W, Xu, H.E, Cherezov, V, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural features for functional selectivity at serotonin receptors.
Science, 340, 2013
|
|
4H33
 
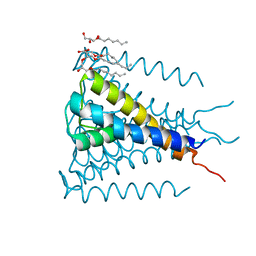 | | Crystal structure of a voltage-gated K+ channel pore module in a closed state in lipid membranes, tetragonal crystal form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lmo2059 protein, POTASSIUM ION | | Authors: | Santos, J.S, Asmar-Rovira, G.A, Han, G.W, Liu, W, Syeda, R, Cherezov, V, Baker, K.A, Stevens, R.C, Montal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Voltage-gated K+ Channel Pore Module in a Closed State in Lipid Membranes.
J.Biol.Chem., 287, 2012
|
|
1F82
 
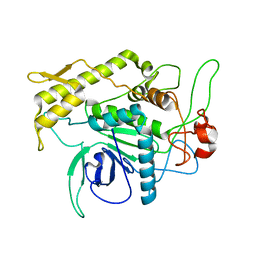 | | BOTULINUM NEUROTOXIN TYPE B CATALYTIC DOMAIN | | Descriptor: | BOTULINUM NEUROTOXIN TYPE B, ZINC ION | | Authors: | Hanson, M.A, Stevens, R.C. | | Deposit date: | 2000-06-28 | | Release date: | 2000-08-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cocrystal structure of synaptobrevin-II bound to botulinum neurotoxin type B at 2.0 A resolution.
Nat.Struct.Biol., 7, 2000
|
|
1GAF
 
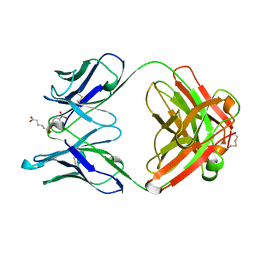 | | 48G7 HYBRIDOMA LINE FAB COMPLEXED WITH HAPTEN 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID | | Descriptor: | 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID, CHIMERIC 48G7 FAB | | Authors: | Wedemayer, G.J, Patten, P.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 1996-02-06 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The immunological evolution of catalysis.
Science, 271, 1996
|
|
1DMW
 
 | | CRYSTAL STRUCTURE OF DOUBLE TRUNCATED HUMAN PHENYLALANINE HYDROXYLASE WITH BOUND 7,8-DIHYDRO-L-BIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, FE (III) ION, PHENYLALANINE HYDROXYLASE | | Authors: | Erlandsen, H, Stevens, R.C, Flatmark, T. | | Deposit date: | 1999-12-15 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and site-specific mutagenesis of pterin-bound human phenylalanine hydroxylase.
Biochemistry, 39, 2000
|
|
1IM1
 
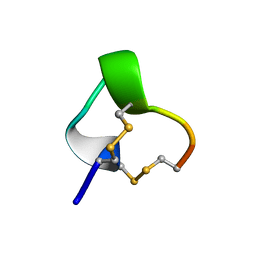 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1, 20 STRUCTURES | | Descriptor: | ALPHA-CONOTOXIN IM1 | | Authors: | Rogers, J.P, Luginbuhl, P, Shen, G.S, Mccabe, R.T, Stevens, R.C, Wemmer, D.E. | | Deposit date: | 1998-11-18 | | Release date: | 1999-06-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-conotoxin ImI and comparison to other conotoxins specific for neuronal nicotinic acetylcholine receptors.
Biochemistry, 38, 1999
|
|
1LTU
 
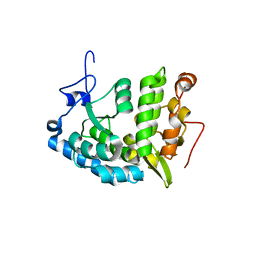 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM, APO (NO IRON BOUND) STRUCTURE | | Descriptor: | PHENYLALANINE-4-HYDROXYLASE | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1LTZ
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE HAS BOUND IRON (III) AND OXIDIZED COFACTOR 7,8-DIHYDROBIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CHLORIDE ION, FE (III) ION, ... | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-21 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1LTV
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE WITH BOUND OXIDIZED Fe(III) | | Descriptor: | FE (III) ION, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
