1PYG
 
 | |
1TRM
 
 | |
3CX8
 
 | |
3CX7
 
 | |
3CX6
 
 | |
1IAP
 
 | |
5DF0
 
 | | Crystal structure of AcMNPV Chitinase A in complex WITH CHITOTRIO-THIAZOLINE DITHIOAMIDE | | Descriptor: | (2R,3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methylhexahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, ... | | Authors: | Mou, T.-C, Sprang, S.R. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Crystal structure of AcMNPV Chitinase A
To Be Published
|
|
5DFT
 
 | |
1TL7
 
 | | Complex Of Gs- With The Catalytic Domains Of Mammalian Adenylyl Cyclase: Complex With 2'(3')-O-(N-methylanthraniloyl)-guanosine 5'-triphosphate and Mn | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase, ... | | Authors: | Mou, T.C, Gille, A, Seifert, R.J, Sprang, S.R. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of mammalian membrane adenylyl cyclase by 2 '(3')-O-(N-Methylanthraniloyl)-guanosine 5 '-triphosphate.
J.Biol.Chem., 280, 2005
|
|
1SVS
 
 | | Structure of the K180P mutant of Gi alpha subunit bound to GppNHp. | | Descriptor: | Guanine nucleotide-binding protein G(i), alpha-1 subunit, MAGNESIUM ION, ... | | Authors: | Thomas, C.J, Du, X, Li, P, Wang, Y, Ross, E.M, Sprang, S.R. | | Deposit date: | 2004-03-29 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uncoupling conformational change from GTP hydrolysis in a heterotrimeric G protein {alpha}-subunit.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SVK
 
 | | Structure of the K180P mutant of Gi alpha subunit bound to AlF4 and GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Thomas, C.J, Du, X, Li, P, Wang, Y, Ross, E.M, Sprang, S.R. | | Deposit date: | 2004-03-29 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncoupling conformational change from GTP hydrolysis in a heterotrimeric G protein {alpha}-subunit.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1NCF
 
 | |
6AU4
 
 | | Crystal structure of the major quadruplex formed in the human c-MYC promoter | | Descriptor: | DNA (5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*A)-3'), POTASSIUM ION | | Authors: | Stump, S, Mou, T.C, Sprang, S.R, Natale, N.R, Beall, H.D. | | Deposit date: | 2017-08-30 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the major quadruplex formed in the promoter region of the human c-MYC oncogene.
PLoS ONE, 13, 2018
|
|
4MU8
 
 | | Crystal structure of an oxidized form of yeast iso-1-cytochrome c at pH 8.8 | | Descriptor: | Cytochrome c iso-1, GLYCEROL, HEME C, ... | | Authors: | McClelland, L.J, Mou, T.-C, Jeakins-Cooley, M.E, Sprang, S.R, Bowler, B.E. | | Deposit date: | 2013-09-20 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a mitochondrial cytochrome c conformer competent for peroxidase activity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6TYL
 
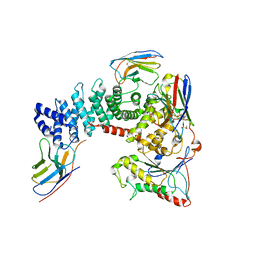 | | Crystal structure of mammalian Ric-8A:Galpha(i):nanobody complex | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1, Nanobody A, Nanobody B, ... | | Authors: | Mou, T.C, McClelland, L, Yates-Hansen, C, Sprang, S.R. | | Deposit date: | 2019-08-09 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the G protein chaperone and guanine nucleotide exchange factor Ric-8A bound to G alpha i1.
Nat Commun, 11, 2020
|
|
6UKT
 
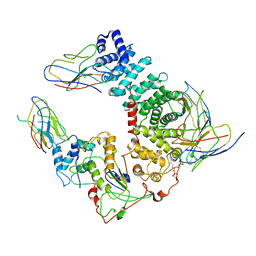 | | Cryo-EM structure of mammalian Ric-8A:Galpha(i):nanobody complex | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1, NB8109, NB8117, ... | | Authors: | Mou, T.C, Zhang, K, Johnston, J.D, Chiu, W, Sprang, S.R. | | Deposit date: | 2019-10-05 | | Release date: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structure of the G protein chaperone and guanine nucleotide exchange factor Ric-8A bound to G alpha i1.
Nat Commun, 11, 2020
|
|
3G82
 
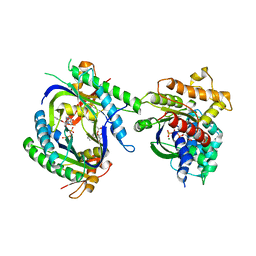 | | Complex of GS-alpha with the catalytic domains of mammalian adenylyl cyclase: complex with MANT-ITP and Mn | | Descriptor: | 3'-O-{[2-(methylamino)phenyl]carbonyl}inosine 5'-(tetrahydrogen triphosphate), 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase type 2, ... | | Authors: | Huebner, M, Mou, T.-C, Sprang, S.R, Seifert, R. | | Deposit date: | 2009-02-11 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | 2',3'-(O)-(N-Methyl)anthraniloyl-inosine 5'-triphosphate is the Most Potent Adenylyl Cyclase 1 and 5 Inhibitor Known so far and Effectively Promotes Catalytic Subunit Assembly in the Absence of Forskolin
To be Published
|
|
3MAA
 
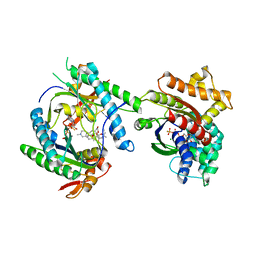 | |
3KZ1
 
 | | Crystal Structure of the Complex of PDZ-RhoGEF DH/PH domains with GTP-gamma-S Activated RhoA | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Rho guanine nucleotide exchange factor 11, ... | | Authors: | Chen, Z, Sternweis, P.C, Sprang, S.R. | | Deposit date: | 2009-12-07 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Activated RhoA binds to the pleckstrin homology (PH) domain of PDZ-RhoGEF, a potential site for autoregulation.
J.Biol.Chem., 285, 2010
|
|
6XAU
 
 | |
1YBT
 
 | | MYCOBACTERIUM TUBERCULOSIS ADENYLYL CYCLASE, RV1900C CHD | | Descriptor: | hydrolase, alpha/beta hydrolase fold family | | Authors: | Sinha, S.C, Wetterer, M, Sprang, S.R, Schultz, J.E, Linder, J.U. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Origin of asymmetry in adenylyl cyclases: structures of Mycobacterium tuberculosis Rv1900c.
Embo J., 24, 2005
|
|
1YBU
 
 | | Mycobacterium tuberculosis adenylyl cyclase Rv1900c CHD, in complex with a substrate analog. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION, lipJ | | Authors: | Sinha, S.C, Wetterer, M, Sprang, S.R, Schultz, J.E, Linder, J.U. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Origin of asymmetry in adenylyl cyclases: structures of Mycobacterium tuberculosis Rv1900c.
Embo J., 24, 2005
|
|
5DEZ
 
 | | Crystal structure of AcMNPV Chitinase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ac-ChiA, ... | | Authors: | Mou, T.-C, Sprang, S.R. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of AcMNPV Chitinase A
To Be Published
|
|
5I56
 
 | | Agonist-bound GluN1/GluN2A agonist binding domains with TCN201 | | Descriptor: | GLUTAMIC ACID, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
1XHM
 
 | | The Crystal Structure of a Biologically Active Peptide (SIGK) Bound to a G Protein Beta:Gamma Heterodimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-2 subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(T) beta subunit 1, SIGK Peptide | | Authors: | Davis, T.L, Bonacci, T.M, Smrcka, A.V, Sprang, S.R. | | Deposit date: | 2004-09-20 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Molecular Characterization of a Preferred Protein Interaction Surface on G Protein betagamma Subunits.
Biochemistry, 44, 2005
|
|
