5D50
 
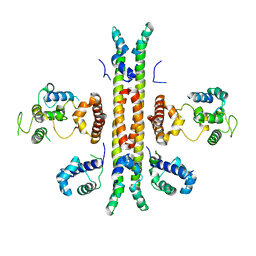 | | Crystal structure of Rep-Ant complex from Salmonella-temperate phage | | Descriptor: | Anti-repressor protein, Repressor | | Authors: | Son, S.H, Yoon, H.J, Ryu, S, Lee, H.H. | | Deposit date: | 2015-08-10 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Noncanonical DNA-binding mode of repressor and its disassembly by antirepressor
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8KEM
 
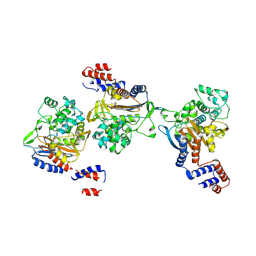 | | PKS domains-fused AmpC EC2 | | Descriptor: | SDR family NAD(P)-dependent oxidoreductase,Beta-lactamase | | Authors: | Son, S.Y, Bae, D.W, Cha, S.S. | | Deposit date: | 2023-08-12 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.76998281 Å) | | Cite: | Structural investigation of the docking domain assembly from trans-AT polyketide synthases.
Structure, 2024
|
|
2Z5Y
 
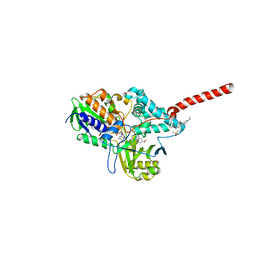 | | Crystal Structure of Human Monoamine Oxidase A (G110A) with Harmine | | Descriptor: | 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Amine oxidase [flavin-containing] A, DECYL(DIMETHYL)PHOSPHINE OXIDE, ... | | Authors: | Son, S.Y, Ma, J, Yoshimura, M, Tsukihara, T. | | Deposit date: | 2007-07-20 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure of human monoamine oxidase A at 2.2-A resolution: The control of opening the entry for substrates/inhibitors
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z5X
 
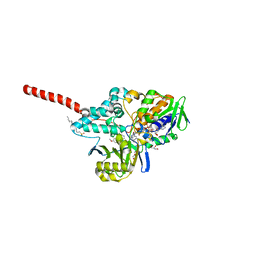 | | Crystal Structure of Human Monoamine Oxidase A with Harmine | | Descriptor: | 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Amine oxidase [flavin-containing] A, DECYL(DIMETHYL)PHOSPHINE OXIDE, ... | | Authors: | Son, S.Y, Ma, J, Yoshimura, M, Tsukihara, T. | | Deposit date: | 2007-07-20 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human monoamine oxidase A at 2.2-A resolution: The control of opening the entry for substrates/inhibitors
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2WGU
 
 | | Structure of human adenovirus serotype 37 fibre head in complex with a sialic acid derivative, O-Methyl 5-N- methoxycarbonyl -3,5-dideoxy- D-glycero-a-D-galacto-2-nonulopyranosylonic acid | | Descriptor: | 3,5-dideoxy-5-[(methoxycarbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, FIBER PROTEIN, ZINC ION | | Authors: | Johansson, S, Nilsson, E, Qian, W, Guilligay, D, Crepin, T, Cusack, S, Arnberg, N, Elofsson, M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Evaluation of N-Acyl Modified Sialic Acids as Inhibitors of Adenoviruses Causing Epidemic Keratoconjunctivitis.
J.Med.Chem., 52, 2009
|
|
2WGT
 
 | | Structure of human adenovirus serotype 37 fibre head in complex with a sialic acid derivative, O-Methyl 5-N-propaonyl-3,5-dideoxy-D- glycero-a-D-galacto-2-nonulopyranosylonic acid | | Descriptor: | 3,5-dideoxy-5-(propanoylamino)-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, FIBER PROTEIN, ZINC ION | | Authors: | Johansson, S, Nilsson, E, Qian, W, Guilligay, D, Crepin, T, Cusack, S, Arnberg, N, Elofsson, M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Evaluation of N-Acyl Modified Sialic Acids as Inhibitors of Adenoviruses Causing Epidemic Keratoconjunctivitis.
J.Med.Chem., 52, 2009
|
|
3W8L
 
 | | Crystal structure of human CK2 in complex with inositol hexakisphosphate | | Descriptor: | Casein kinase II subunit alpha, INOSITOL HEXAKISPHOSPHATE | | Authors: | Son, S.H, Lee, W.-K, Yu, Y.G, Lee, H.H. | | Deposit date: | 2013-03-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ENJ
 
 | | Structure of Pig Heart Citrate Synthase at 1.78 A resolution | | Descriptor: | CHLORIDE ION, CYSTEINE, Citrate synthase, ... | | Authors: | Larson, S.B, Day, J.S, Nguyen, C, Cudney, R, McPherson, A, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2008-09-25 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of pig heart citrate synthase at 1.78 A resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3EDN
 
 | | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family | | Descriptor: | MAGNESIUM ION, Phenazine biosynthesis protein, PhzF family, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-03 | | Release date: | 2008-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family
To be Published
|
|
7AKV
 
 | | The cryo-EM structure of the Vag8-C1 inhibitor complex | | Descriptor: | Plasma protease C1 inhibitor, Vag8 | | Authors: | Johnson, S, Lea, S.M, Deme, J.C, Furlong, E, Dhillon, A. | | Deposit date: | 2020-10-02 | | Release date: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular Basis for Bordetella pertussis Interference with Complement, Coagulation, Fibrinolytic, and Contact Activation Systems: the Cryo-EM Structure of the Vag8-C1 Inhibitor Complex.
Mbio, 12, 2021
|
|
8KCN
 
 | |
2J0O
 
 | | Shigella Flexneri IpaD | | Descriptor: | GLYCEROL, INVASIN IPAD | | Authors: | Johnson, S, Roversi, P, Lea, S.M. | | Deposit date: | 2006-08-04 | | Release date: | 2006-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD.
J. Biol. Chem., 282, 2007
|
|
2J5A
 
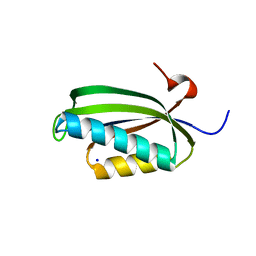 | | Folding of S6 structures with divergent amino-acid composition: pathway flexibility within partly overlapping foldons | | Descriptor: | 30S RIBOSOMAL PROTEIN S6, SODIUM ION | | Authors: | Hansson, S, Olofsson, L, Hedberg, L, Oliveberg, M, Logan, D.T. | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Folding of S6 Structures with Divergent Amino Acid Composition: Pathway Flexibility within Partly Overlapping Foldons.
J.Mol.Biol., 365, 2007
|
|
1SZJ
 
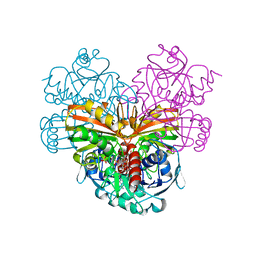 | |
1JNU
 
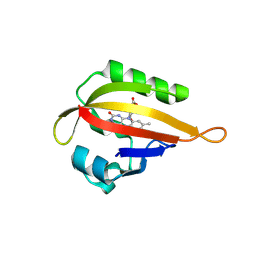 | |
6PNQ
 
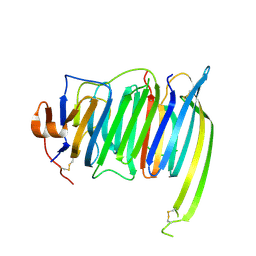 | | Crystal structure of the SS2A splice insert-containing neurexin-1 LNS2 domain in complex with neurexophilin-1 | | Descriptor: | Neurexin-1, Neurexophilin-1 | | Authors: | Wilson, S.C, White, K.I, Zhou, Q, Brunger, A.T. | | Deposit date: | 2019-07-02 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Structures of neurexophilin-neurexin complexes reveal a regulatory mechanism of alternative splicing.
Embo J., 38, 2019
|
|
5OMU
 
 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5OMR
 
 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with vanillin. | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, GcoA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5OMS
 
 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with guaethol. | | Descriptor: | 2-ethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5OGX
 
 | | Crystal structure of Amycolatopsis cytochrome P450 reductase GcoB. | | Descriptor: | BROMIDE ION, Cytochrome P450 reductase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-07-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5NCB
 
 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with guaiacol. | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-07-04 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5NQP
 
 | | Structure of a fHbp(V1.4):PorA(P1.16) chimera. Fusion at fHbp position 151. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Factor H binding protein variant B16_001,Major outer membrane protein P.IA,Factor H binding protein variant B16_001, ... | | Authors: | Johnson, S, Hollingshead, S, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
5NQZ
 
 | | Structure of a fHbp(V1.1):PorA(P1.16) chimera. Fusion at fHbp position 309. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Factor H binding protein,Major outer membrane protein P.IA,Factor H binding protein, ... | | Authors: | Johnson, S, Hollingshead, S, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
5NQY
 
 | | Structure of a fHbp(V1.4):PorA(P1.16) chimera. Fusion at fHbp position 309. | | Descriptor: | Factor H binding protein variant B16_001,Major outer membrane protein P.IA,Factor H binding protein variant B16_001 | | Authors: | Johnson, S, Hollingshead, S, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
5NQX
 
 | | Structure of a fHbp(V1.1):PorA(P1.16) chimera. Fusion at fHbp position 294. | | Descriptor: | Factor H binding protein,Major outer membrane protein P.IA,Factor H binding protein | | Authors: | Johnson, S, Jongerius, I, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
