4ZQ3
 
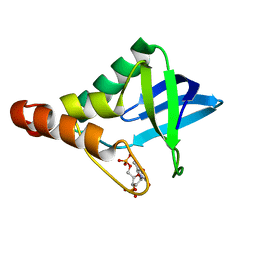 | |
4ZUI
 
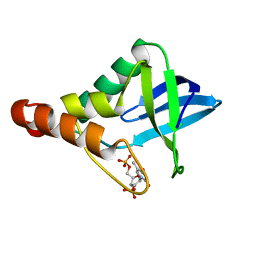 | |
6MBE
 
 | | Human Mcl-1 in complex with the designed peptide dM7 | | Descriptor: | CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1, dM7 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
8AF2
 
 | | Human Sterol Carrier Protein with unnatural amino acid 2,2'-bipyridine alanine incorporated at position 111 | | Descriptor: | COPPER (II) ION, Enoyl-CoA hydratase 2, FRAGMENT OF TRITON X-100, ... | | Authors: | Richardson, J.M, Klemencic, E, Jarvis, A.G. | | Deposit date: | 2022-07-15 | | Release date: | 2023-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Using BpyAla to generate copper artificial metalloenzymes: a catalytic and structural study.
Catalysis Science And Technology, 14, 2024
|
|
6MBC
 
 | | Human Bfl-1 in complex with the designed peptide dF4 | | Descriptor: | Bcl-2-related protein A1, dF4 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6MBD
 
 | | Human Mcl-1 in complex with the designed peptide dM1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, dM1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6UVJ
 
 | |
6UWX
 
 | |
6UVM
 
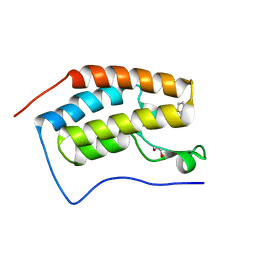 | | Cocrystal of BRD4(D1) with a methyl carbamate thiazepane inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-[(7S)-7-(thiophen-2-yl)-6,7-dihydro-1,4-thiazepin-4(5H)-yl]ethan-1-one, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Pomerantz, W.C.K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Evaluating the Advantages of Using 3D-Enriched Fragments for Targeting BET Bromodomains.
Acs Med.Chem.Lett., 10, 2019
|
|
3NMO
 
 | |
8GER
 
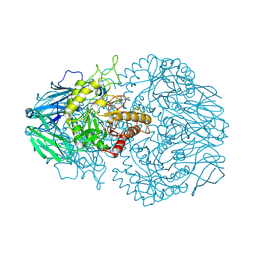 | | E. eligens beta-glucuronidase bound to norquetiapine-glucuronide | | Descriptor: | 11-(4-beta-D-glucopyranuronosylpiperazin-1-yl)dibenzo[b,f][1,4]thiazepine, Beta-glucuronidase | | Authors: | Simpson, J.B, Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8GEO
 
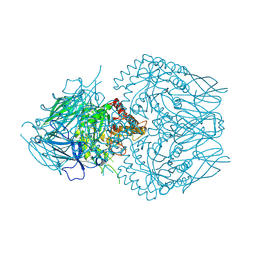 | | E. eligens beta-glucuronidase bound to 3-OH-desloratidine-glucuronide | | Descriptor: | 8-chloro-11-(1-beta-D-glucopyranuronosylpiperidin-4-ylidene)-3-hydroxy-6,11-dihydro-5H-benzo[5,6]cyclohepta[1,2-b]pyridine, Beta-glucuronidase, GLYCEROL | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8GET
 
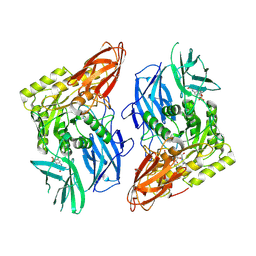 | | R. hominis 2 beta-glucuronidase bound to norquetiapine-glucuronide | | Descriptor: | 11-(4-beta-D-glucopyranuronosylpiperazin-1-yl)dibenzo[b,f][1,4]thiazepine, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8GEN
 
 | | E. eligens beta-glucuronidase bound to UNC10201652-glucuronide | | Descriptor: | 8-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5-(morpholin-4-yl)-1,2,3,4-tetrahydro[1,2,3]triazino[4',5':4,5]thieno[2,3 -c]isoquinoline, Beta-glucuronidase | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8GES
 
 | | R. hominis 2 beta-glucuronidase bound to UNC10201652-glucuronide | | Descriptor: | 8-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5-(morpholin-4-yl)-1,2,3,4-tetrahydro[1,2,3]triazino[4',5':4,5]thieno[2,3 -c]isoquinoline, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8GEQ
 
 | | E. eligens beta-glucuronidase bound to ceritinib-glucuronide | | Descriptor: | 4-amino-5-chloro-2-{4-(1-beta-D-glucopyranuronosylpiperidin-4-yl)-5-methyl-2-[(propan-2-yl)oxy]anilino}pyrimidine, Beta-glucuronidase | | Authors: | Simpson, J.B, Kowalewski, M.K, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
2KGI
 
 | |
2M0O
 
 | |
2KU7
 
 | |
8AF3
 
 | | Sterol carrier protein Artifical metalloenzyme incorporating Q111C mutation coupled to 2,2'-bipyridine | | Descriptor: | COPPER (II) ION, Enoyl-CoA hydratase 2, FRAGMENT OF TRITON X-100, ... | | Authors: | Richardson, J.M, Klemencic, E, Jarvis, A.G. | | Deposit date: | 2022-07-15 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Using BpyAla to generate copper artificial metalloenzymes: a catalytic and structural study.
Catalysis Science And Technology, 14, 2024
|
|
6BJZ
 
 | |
1R7I
 
 | | HMG-CoA Reductase from P. mevalonii, native structure at 2.2 angstroms resolution. | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, SULFATE ION | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-10-21 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-COA Reductase.
To be Published
|
|
1R31
 
 | | HMG-CoA reductase from Pseudomonas mevalonii complexed with HMG-CoA | | Descriptor: | (R)-MEVALONATE, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, COENZYME A, ... | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-CoA Reductase.
To be Published
|
|
2X3H
 
 | | COLIPHAGE K5A LYASE | | Descriptor: | BROMIDE ION, K5 LYASE | | Authors: | Thompson, J.E, Pourhossein, M, Goldrick, M, Hudson, T, Derrick, J.P, Roberts, I.S. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The K5 Lyase Kfla Combines a Viral Tail Spike Structure with a Bacterial Polysaccharide Lyase Mechanism.
J.Biol.Chem., 285, 2010
|
|
1H3I
 
 | | Crystal structure of the Histone Methyltransferase SET7/9 | | Descriptor: | HISTONE H3 LYSINE 4 SPECIFIC METHYLTRANSFERASE, MAGNESIUM ION | | Authors: | Wilson, J.R, Jing, C, Walker, P.A, Martin, S.R, Howell, S.A, Blackburn, G.M, Gamblin, S.J, Xiao, B. | | Deposit date: | 2002-09-04 | | Release date: | 2002-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Functional Analysis of the Histone Methyltransferase Set7/9
Cell(Cambridge,Mass.), 111, 2002
|
|
