1C0E
 
 | | Active Site S19A Mutant of Bovine Heart Phosphotyrosyl Phosphatase | | Descriptor: | PHOSPHATE ION, PROTEIN (TYROSINE PHOSPHATASE (ORTHOPHOSPHORIC MONOESTER PHOSPHOHYDROLASE)) | | Authors: | Tabernero, L, Evans, B.N, Tishmack, P.A, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 1999-07-15 | | Release date: | 1999-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the bovine protein tyrosine phosphatase dimer reveals a potential self-regulation mechanism.
Biochemistry, 38, 1999
|
|
121D
 
 | | MOLECULAR STRUCTURE OF THE A-TRACT DNA DODECAMER D(CGCAAATTTGCG) COMPLEXED WITH THE MINOR GROOVE BINDING DRUG NETROPSIN | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3'), NETROPSIN | | Authors: | Tabernero, L, Verdaguer, N, Coll, M, Fita, I, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Aymami, J. | | Deposit date: | 1993-04-14 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor groove binding drug netropsin.
Biochemistry, 32, 1993
|
|
1AHR
 
 | |
1T02
 
 | | Crystal structure of a Statin bound to class II HMG-CoA reductase | | Descriptor: | (3R,5R)-7-((1R,2R,6S,8R,8AS)-2,6-DIMETHYL-8-{[(2R)-2-METHYLBUTANOYL]OXY}-1,2,6,7,8,8A-HEXAHYDRONAPHTHALEN-1-YL)-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Tabernero, L, Rodwell, V.W, Stauffacher, C. | | Deposit date: | 2004-04-07 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a statin bound to a class II hydroxymethylglutaryl-CoA reductase.
J.Biol.Chem., 278, 2003
|
|
1HDT
 
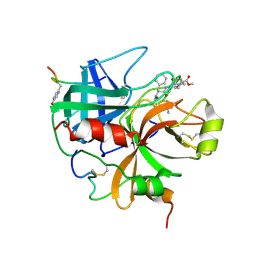 | |
1QAY
 
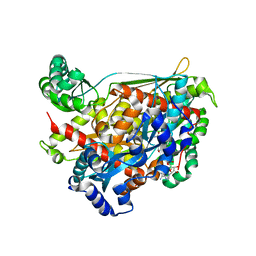 | | TERNARY COMPLEX OF PSEUDOMONAS MEVALONII HMG-COA REDUCTASE WITH MEVALONATE AND NAD+ | | Descriptor: | (R)-MEVALONATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (3-HYDROXY-3-METHYLGLUTARYL-COENZYME A REDUCTASE) | | Authors: | Tabernero, L, Bochar, D.A, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 1999-04-07 | | Release date: | 1999-06-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate-induced closure of the flap domain in the ternary complex structures provides insights into the mechanism of catalysis by 3-hydroxy-3-methylglutaryl-CoA reductase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QAX
 
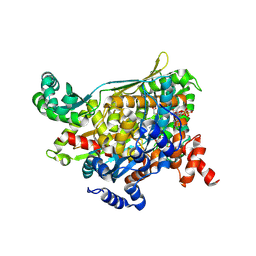 | | TERNARY COMPLEX OF PSEUDOMONAS MEVALONII HMG-COA REDUCTASE WITH HMG-COA AND NAD+ | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (3-HYDROXY-3-METHYLGLUTARYL-COENZYME A REDUCTASE) | | Authors: | Tabernero, L, Bochar, D.A, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 1999-04-06 | | Release date: | 1999-06-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate-induced closure of the flap domain in the ternary complex structures provides insights into the mechanism of catalysis by 3-hydroxy-3-methylglutaryl-CoA reductase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2ZMC
 
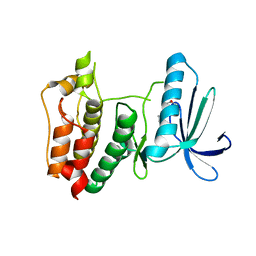 | | Crystal structure of human mitotic checkpoint kinase Mps1 catalytic domain apo form | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK | | Authors: | Chu, M.L.H, Chavas, L.M.G, Douglas, K.T, Eyers, P.A, Tabernero, L. | | Deposit date: | 2008-04-16 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of the catalytic domain of the mitotic checkpoint kinase Mps1 in complex with SP600125.
J.Biol.Chem., 283, 2008
|
|
2ZMD
 
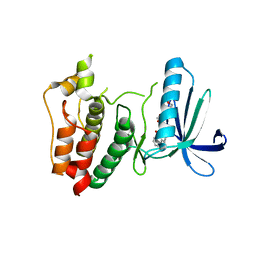 | | Crystal structure of human Mps1 catalytic domain T686A mutant in complex with SP600125 inhibitor | | Descriptor: | 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK | | Authors: | Chu, M.L.H, Chavas, L.M.G, Douglas, K.T, Eyers, P.A, Tabernero, L. | | Deposit date: | 2008-04-16 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the catalytic domain of the mitotic checkpoint kinase Mps1 in complex with SP600125.
J.Biol.Chem., 283, 2008
|
|
3HMN
 
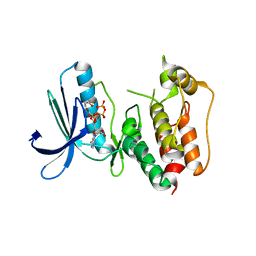 | | Crystal structure of human Mps1 catalytic domain in complex with ATP | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity protein kinase TTK | | Authors: | Chu, M.L.H, Chavas, L.M.G, Williams, D.H, Tabernero, L, Eyers, P.A. | | Deposit date: | 2009-05-29 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biophysical and X-ray crystallographic analysis of Mps1 kinase inhibitor complexes.
Biochemistry, 49, 2010
|
|
3HMO
 
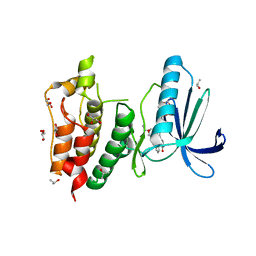 | | Crystal structure of human Mps1 catalytic domain in complex with the inhibitor staurosporine | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK, GLYCEROL, ... | | Authors: | Chu, M.L.H, Chavas, L.M.G, Williams, D.H, Tabernero, L, Eyers, P.A. | | Deposit date: | 2009-05-29 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biophysical and X-ray crystallographic analysis of Mps1 kinase inhibitor complexes.
Biochemistry, 49, 2010
|
|
3HMP
 
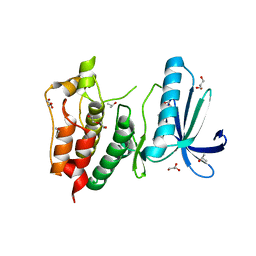 | | Crystal structure of human Mps1 catalytic domain in complex with a quinazolin ligand Compound 4 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 7-chloro-N-(cyclopropylmethyl)quinazolin-4-amine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chu, M.L.H, Chavas, L.M.G, Williams, D.H, Tabernero, L, Eyers, P.A. | | Deposit date: | 2009-05-29 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biophysical and X-ray crystallographic analysis of Mps1 kinase inhibitor complexes.
Biochemistry, 49, 2010
|
|
1D1P
 
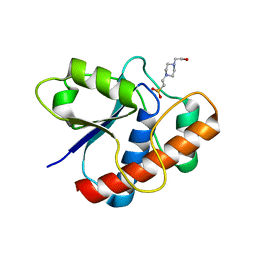 | | CRYSTAL STRUCTURE OF A YEAST LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE (LTP1) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TYROSINE PHOSPHATASE | | Authors: | Wang, S, Tabernero, L, Zhang, M, Harms, E, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 1999-09-20 | | Release date: | 2000-03-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate.
Biochemistry, 39, 2000
|
|
1D1Q
 
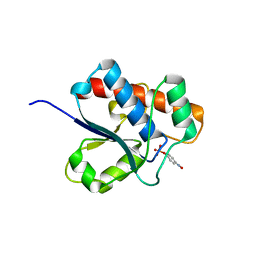 | | CRYSTAL STRUCTURE OF A YEAST LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE (LTP1) COMPLEXED WITH THE SUBSTRATE PNPP | | Descriptor: | 4-NITROPHENYL PHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Wang, S, Tabernero, L, Zhang, M, Harms, E, Van Etten, R.L, Staufacher, C.V. | | Deposit date: | 1999-09-20 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate.
Biochemistry, 39, 2000
|
|
2UXS
 
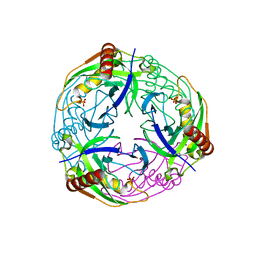 | | 2.7A crystal structure of inorganic pyrophosphatase (Rv3628) from Mycobacterium tuberculosis at pH 7.5 | | Descriptor: | INORGANIC PYROPHOSPHATASE, PHOSPHATE ION | | Authors: | Cole, R.E, Cianci, M, Hall, J.F, Matsuda, T, Kigawa, T, Yokoyama, S, Hasnain, S.S, Tabernero, L. | | Deposit date: | 2007-03-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Rv3628: An Inorganic Pyrophosphatase from Mycobacterium Tuberculosis
To be Published
|
|
1R7I
 
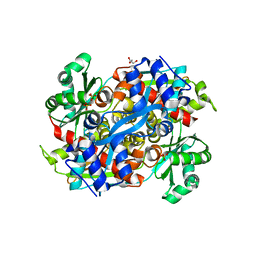 | | HMG-CoA Reductase from P. mevalonii, native structure at 2.2 angstroms resolution. | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, SULFATE ION | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-10-21 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-COA Reductase.
To be Published
|
|
1R31
 
 | | HMG-CoA reductase from Pseudomonas mevalonii complexed with HMG-CoA | | Descriptor: | (R)-MEVALONATE, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, COENZYME A, ... | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-CoA Reductase.
To be Published
|
|
