8C8S
 
 | | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 21). | | Descriptor: | (2~{R})-3-[6-chloranyl-2-(prop-2-enylamino)quinazolin-4-yl]-2-methyl-~{N}-oxidanyl-propanamide, DNA cross-link repair 1A protein, ZINC ION | | Authors: | Yosaatmadja, Y, Newman, J.A, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-01-20 | | Release date: | 2024-01-31 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
8C8B
 
 | | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 48). | | Descriptor: | 4-[[(2~{S})-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-~{N}-prop-2-enyl-quinazoline-2-carboxamide, DNA cross-link repair 1A protein, ZINC ION | | Authors: | Yosaatmadja, Y, Newman, J.A, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
4JAA
 
 | | Factor inhibiting HIF-1 alpha in complex with consensus ankyrin repeat domain-(d)LEU peptide | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-(d)LEU, Hypoxia-inducible factor 1-alpha inhibitor, N-OXALYLGLYCINE, ... | | Authors: | Scotti, J.S, Ge, W, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-02-18 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of an oxygenase in complex with substrate
To be Published
|
|
8C8D
 
 | | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 44). | | Descriptor: | (2~{R})-3-[6-chloranyl-2-(furan-2-ylmethylamino)quinazolin-4-yl]-2-methyl-~{N}-oxidanyl-propanamide, DNA cross-link repair 1A protein, ZINC ION | | Authors: | Yosaatmadja, Y, Newman, J.A, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
8CEW
 
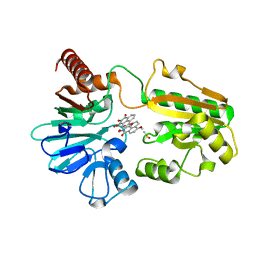 | | Crystal structure of human DNA cross-link repair 1A in complex with N-hydroxyimide inhibitor H1 | | Descriptor: | 6-methoxy-2-oxidanyl-benzo[de]isoquinoline-1,3-dione, DIMETHYL SULFOXIDE, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
8CF0
 
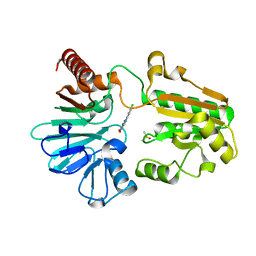 | | Crystal structure of human DNA cross-link repair 1A in complex with quinoxalinedione inhibitor H2 | | Descriptor: | 9-chloranyl-1,4-dihydropyrazino[2,3-c]quinoline-2,3-dione, DIMETHYL SULFOXIDE, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
8CG9
 
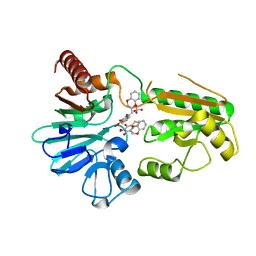 | | Crystal structure of human DNA cross-link repair 1A in complex with a cyclic N-hydroxyurea inhibitor | | Descriptor: | 1-[(2S)-2,3-dihydro-1,4-benzodioxin-2-ylmethyl]-3-hydroxythieno[3,2-d]pyrimidine-2,4(1H,3H)-dione, 1-[[(3~{R})-2,3-dihydro-1,4-benzodioxin-3-yl]methyl]-3-oxidanyl-thieno[3,2-d]pyrimidine-2,4-dione, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
4JHT
 
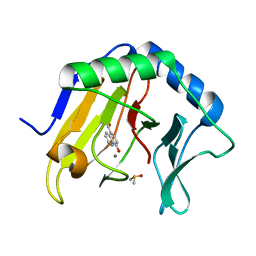 | | Crystal Structure of AlkB in complex with 5-carboxy-8-hydroxyquinoline (IOX1) | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, ACETATE ION, Alpha-ketoglutarate-dependent dioxygenase AlkB, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-03-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 5-Carboxy-8-hydroxyquinoline is a Broad Spectrum 2-Oxoglutarate Oxygenase Inhibitor which Causes Iron Translocation.
Chem Sci, 4, 2013
|
|
4IE0
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with pyridine-2,4-dicarboxylate (2,4-PDCA) | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, PYRIDINE-2,4-DICARBOXYLIC ACID, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
4NHM
 
 | | Crystal structure of Tpa1p from Saccharomyces cerevisiae, termination and polyadenylation protein 1, in complex with N-[(1-chloro-4-hydroxyisoquinolin-3-yl)carbonyl]glycine (IOX3/UN9) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE, ... | | Authors: | Scotti, J.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Ribosomal Oxygenase OGFOD1 Provides Insights into the Regio- and Stereoselectivity of Prolyl Hydroxylases.
Structure, 23, 2015
|
|
4NHX
 
 | | Crystal structure of human OGFOD1, 2-oxoglutarate and iron-dependent oxygenase domain containing 1, in complex with N-oxalylglycine (NOG) | | Descriptor: | 2-oxoglutarate and iron-dependent oxygenase domain-containing protein 1, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Horita, S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structure of the Ribosomal Oxygenase OGFOD1 Provides Insights into the Regio- and Stereoselectivity of Prolyl Hydroxylases.
Structure, 23, 2015
|
|
4NJ4
 
 | | Crystal Structure of Human ALKBH5 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation.
Nucleic Acids Res., 42, 2014
|
|
4NHK
 
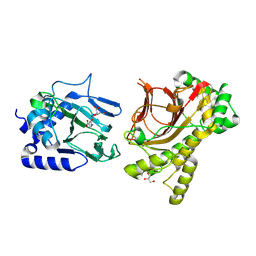 | | Crystal structure of Tpa1p from Saccharomyces cerevisiae, termination and polyadenylation protein 1, in complex with pyridine-2,4-dicarboxylic acid (2,4-PDCA) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PKHD-type hydroxylase TPA1, ... | | Authors: | Scotti, J.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Ribosomal Oxygenase OGFOD1 Provides Insights into the Regio- and Stereoselectivity of Prolyl Hydroxylases.
Structure, 23, 2015
|
|
4NHY
 
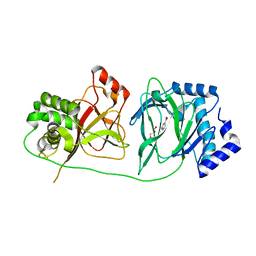 | | Crystal structure of human OGFOD1, 2-oxoglutarate and iron-dependent oxygenase domain containing 1, in complex with pyridine-2,4-dicarboxylic acid (2,4-PDCA) | | Descriptor: | 2-oxoglutarate and iron-dependent oxygenase domain-containing protein 1, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Horita, S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structure of the Ribosomal Oxygenase OGFOD1 Provides Insights into the Regio- and Stereoselectivity of Prolyl Hydroxylases.
Structure, 23, 2015
|
|
4PVT
 
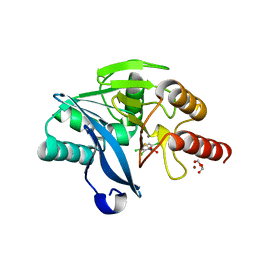 | | Crystal Structure of VIM-2 metallo-beta-lactamase in complex with ML302F | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Aik, W.S, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rhodanine hydrolysis leads to potent thioenolate mediated metallo-beta-lactamase inhibition.
Nat.Chem., 6, 2014
|
|
4PVO
 
 | | Crystal Structure of VIM-2 metallo-beta-lactamase in complex with ML302 and ML302F | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase class B VIM-2, DIMETHYL SULFOXIDE, ... | | Authors: | Aik, W.S, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Rhodanine hydrolysis leads to potent thioenolate mediated metallo-beta-lactamase inhibition.
Nat.Chem., 6, 2014
|
|
4NHL
 
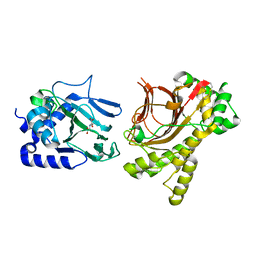 | | Crystal structure of Tpa1p from Saccharomyces cerevisiae, termination and polyadenylation protein 1, in complex with N-oxalylglycine (NOG) | | Descriptor: | MANGANESE (II) ION, N-OXALYLGLYCINE, PKHD-type hydroxylase TPA1 | | Authors: | Scotti, J.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of the Ribosomal Oxygenase OGFOD1 Provides Insights into the Regio- and Stereoselectivity of Prolyl Hydroxylases.
Structure, 23, 2015
|
|
4QHO
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with CCO10 | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, N-{[3-hydroxy-6-(naphthalen-1-yl)pyridin-2-yl]carbonyl}glycine, ZINC ION | | Authors: | Aik, W.S, Clunie-O'Connor, C, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2014-05-28 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors.
J.Med.Chem., 64, 2021
|
|
6RBJ
 
 | | Crystal structure of KDM3B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol | | Descriptor: | 1,2-ETHANEDIOL, 5-(1~{H}-1,2,3,4-tetrazol-5-yl)quinolin-8-ol, CHLORIDE ION, ... | | Authors: | Johansson, C, Newman, J.A, Kawamura, A, Schofield, C.J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-04-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Crystal structure of KDM3B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol
To Be Published
|
|
4BU2
 
 | | 60S ribosomal protein L27A histidine hydroxylase (MINA53) in complex with Ni(II) and 2-oxoglutarate (2OG) | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE MINA, ... | | Authors: | Chowdhury, R, Clifton, I.J, McDonough, M.A, Ng, S.S, Pilka, E, Oppermann, U, Schofield, C.J. | | Deposit date: | 2013-06-19 | | Release date: | 2014-05-14 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
6RZ0
 
 | |
6RMF
 
 | | Crystal structure of NDM-1 with VNRX-5133 | | Descriptor: | (10aR)-2-(((1r,4R)-4-((2-aminoethyl)amino)cyclohexyl)methyl)-6-carboxy-4-hydroxy-4,10a-dihydro-10H-benzo[5,6][1,2]oxaborinino[2,3-b][1,4,2]oxazaborol-4-uide, (4~{R})-4-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, Metallo-beta-lactamase type 2, ... | | Authors: | Hinchliffe, P, Spencer, J, Brem, J, Schofield, C.J. | | Deposit date: | 2019-05-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Bicyclic Boronate VNRX-5133 Inhibits Metallo- and Serine-beta-Lactamases.
J.Med.Chem., 62, 2019
|
|
6RPN
 
 | | Structure of metallo beta lactamase VIM-2 with cyclic boronate APC308. | | Descriptor: | (3~{R})-2,2-bis(oxidanyl)-3-(phenylmethylsulfanyl)-3,4-dihydro-1,2-benzoxaborinin-2-ium-8-carboxylic acid, (3~{S})-2,2-bis(oxidanyl)-3-(phenylmethylsulfanyl)-3,4-dihydro-1,2-benzoxaborinin-2-ium-8-carboxylic acid, Beta-lactamase VIM-2, ... | | Authors: | Parkova, A, Lucic, A, Brem, J, McDonough, M.A, Langley, G.W, Schofield, C.J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Broad Spectrum beta-Lactamase Inhibition by a Thioether Substituted Bicyclic Boronate.
Acs Infect Dis., 6, 2020
|
|
4CCK
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in complex with Mn(II) and N-oxalylglycine (NOG) | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCN
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66 L299C/C300S) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G220C) peptide fragment (complex-2) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
