2PSU
 
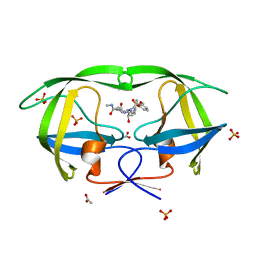 | | Crystal Structure of wild type HIV-1 protease in complex with CARB-AD37 | | Descriptor: | ACETATE ION, N-[(1S,2R)-1-BENZYL-3-{(CYCLOPROPYLMETHYL)[(3-METHOXYPHENYL)SULFONYL]AMINO}-2-HYDROXYPROPYL]-N'-METHYLSUCCINAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-05-07 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design of Mutation-resistant HIV Protease Inhibitors with the Substrate Envelope Hypothesis.
Chem.Biol.Drug Des., 69, 2007
|
|
2Q55
 
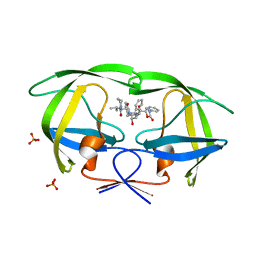 | | Crystal structure of KK44 bound to HIV-1 protease | | Descriptor: | (5S)-N-[(1S,2S,4S)-1-BENZYL-2-HYDROXY-4-{[(2S)-3-METHYL-2-(2-OXOTETRAHYDROPYRIMIDIN-1(2H)-YL)BUTANOYL]AMINO}-5-PHENYLPENTYL]-2-OXO-3-PHENYL-1,3-OXAZOLIDINE-5-CARBOXAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-05-31 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of HIV-1 Protease Inhibitors Incorporating Oxazolidinones as P2/P2' Ligands in Pseudosymmetric Dipeptide Isosteres.
J.Med.Chem., 50, 2007
|
|
2Q3K
 
 | |
2PSV
 
 | |
2Q5K
 
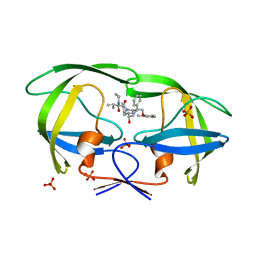 | | Crystal structure of lopinavir bound to wild type HIV-1 protease | | Descriptor: | N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-06-01 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and Synthesis of HIV-1 Protease Inhibitors Incorporating Oxazolidinones as P2/P2' Ligands in Pseudosymmetric Dipeptide Isosteres.
J.Med.Chem., 50, 2007
|
|
2Q54
 
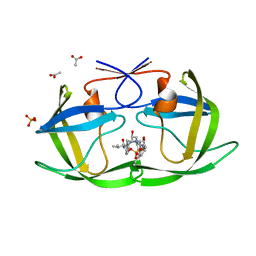 | | Crystal structure of KB73 bound to HIV-1 protease | | Descriptor: | ACETATE ION, N~2~-ACETYL-N-[(1S,3S,4S)-4-({[(5S)-3-(3-ACETYLPHENYL)-2-OXO-1,3-OXAZOLIDIN-5-YL]CARBONYL}AMINO)-1-BENZYL-3-HYDROXY-5-PHENYLPENTYL]-L-VALINAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-05-31 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Synthesis of HIV-1 Protease Inhibitors Incorporating Oxazolidinones as P2/P2' Ligands in Pseudosymmetric Dipeptide Isosteres.
J.Med.Chem., 50, 2007
|
|
2QHZ
 
 | | Crystal structure of protease inhibitor, MIT-1-AC87 in complex with wild type HIV-1 protease | | Descriptor: | (2E)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{(2-THIENYLMETHYL)[(2,4,5-TRIFLUOROPHENYL)SULFONYL]AMINO}PROPYL]-4,4,4-TRIFLUORO-3-METHYLBUT-2-ENAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QHY
 
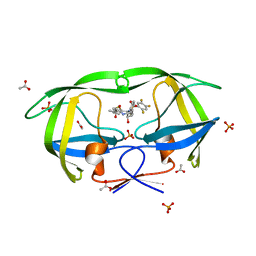 | | Crystal Structure of protease inhibitor, MIT-1-AC86 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N~2~-ACETYL-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{(2-THIENYLMETHYL)[(2,4,5-TRIFLUOROPHENYL)SULFONYL]AMINO}PROPYL]-L-ALANINAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
1KJF
 
 | |
1KJ4
 
 | |
1KJ7
 
 | |
1KJG
 
 | |
4F75
 
 | |
3OXX
 
 | | Crystal Structure of HIV-1 I50V, A71V Protease in Complex with the Protease Inhibitor Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, 1,2-ETHANEDIOL, ... | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-09-22 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50.
J.Virol., 87, 2013
|
|
1KJH
 
 | |
4F73
 
 | |
4F76
 
 | |
3RC5
 
 | |
3RC6
 
 | |
4DJQ
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with MKP86 | | Descriptor: | 2-(2-oxoimidazolidin-1-yl)ethyl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, PHOSPHATE ION, Pol polyprotein | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2012-02-02 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis, and biological and structural evaluations of novel HIV-1 protease inhibitors to combat drug resistance.
J.Med.Chem., 55, 2012
|
|
4DQG
 
 | |
3SU4
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease,NS4A protein, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SV8
 
 | | Crystal structure of NS3/4A protease variant D168A in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SV9
 
 | | Crystal structure of NS3/4A protease variant A156T in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SUE
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
