5MZ6
 
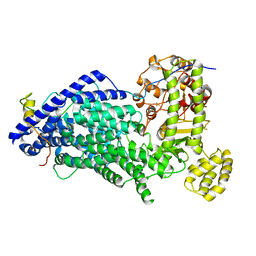 | | Cryo-EM structure of a Separase-Securin complex from Caenorhabditis elegans at 3.8 A resolution | | Descriptor: | Interactor of FizzY protein, SEParase | | Authors: | Boland, A, Martin, T.G, Zhang, Z, Yang, J, Bai, X.C, Chang, L, Scheres, S.H.W, Barford, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-08 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6QJP
 
 | | Cryo-EM structure of heparin-induced 2N4R tau jagged filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6QJH
 
 | | Cryo-EM structure of heparin-induced 2N4R tau snake filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6QJM
 
 | | Cryo-EM structure of heparin-induced 2N4R tau twister filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6QJQ
 
 | | Cryo-EM structure of heparin-induced 2N3R tau filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6RZA
 
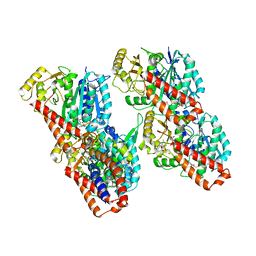 | | Cryo-EM structure of the human inner arm dynein DNAH7 microtubule binding domain bound to microtubules | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1,Dynein heavy chain 7, axonemal,Cytoplasmic dynein 1 heavy chain 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lacey, S.E, He, S, Scheres, S.H.W, Carter, A.P. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-17 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Cryo-EM of dynein microtubule-binding domains shows how an axonemal dynein distorts the microtubule.
Elife, 8, 2019
|
|
6RZB
 
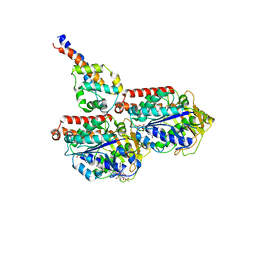 | | Cryo-EM structure of mouse cytoplasmic dynein-1 microtubule binding domain bound to microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lacey, S.E, He, S, Scheres, S.H.W, Carter, A.P. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-17 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Cryo-EM of dynein microtubule-binding domains shows how an axonemal dynein distorts the microtubule.
Elife, 8, 2019
|
|
6ZOZ
 
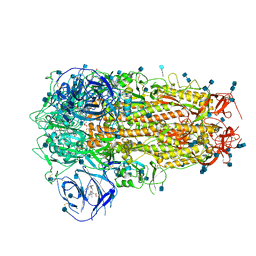 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP0
 
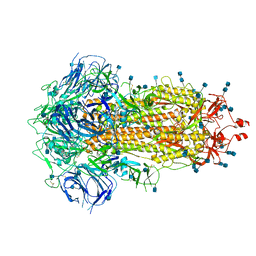 | | Structure of SARS-CoV-2 Spike Protein Trimer (single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP1
 
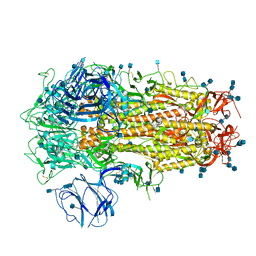 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7A4M
 
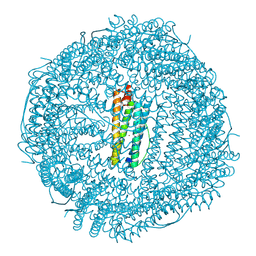 | | Cryo-EM structure of mouse heavy-chain apoferritin at 1.22 A | | Descriptor: | FE (III) ION, Ferritin heavy chain, ZINC ION | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-06-21 | | Method: | ELECTRON MICROSCOPY (1.22 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
7A5V
 
 | | CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-22 | | Release date: | 2020-11-18 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
6ZOX
 
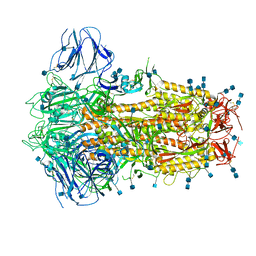 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x2 disulphide-bond mutant, G413C, V987C, single Arg S1/S2 cleavage site) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP2
 
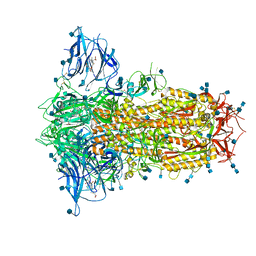 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZWV
 
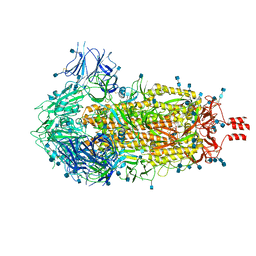 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: 3 Closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ke, Z, Qu, K, Nakane, T, Xiong, X, Cortese, M, Zila, V, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-28 | | Release date: | 2020-08-05 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and distributions of SARS-CoV-2 spike proteins on intact virions.
Nature, 588, 2020
|
|
6ZOY
 
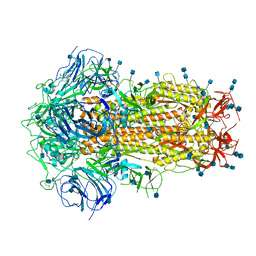 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7ARE
 
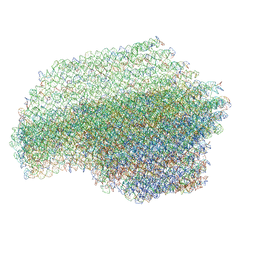 | | DNA origami pointer object v2 | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Thomas, M, Feigl, E, Kohler, F, Kube, M, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7AS5
 
 | | 126 helix bundle DNA nanostructure | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7ART
 
 | | 48 helix bundle DNA origami brick | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Feigl, E, Kube, M, Kohler, F, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7ARV
 
 | | TwistTower_native-twist | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7ARY
 
 | | Twist-Tower_twist-corrected-variant | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
3JBT
 
 | | Atomic structure of the Apaf-1 apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Zhou, M, Li, Y, Hu, Q, Bai, X, Huang, W, Yan, C, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1
Genes Dev., 29, 2015
|
|
5FKU
 
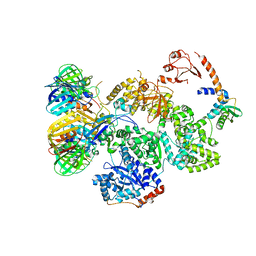 | | cryo-EM structure of the E. coli replicative DNA polymerase complex in DNA free state (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III SUBUNIT ALPHA, DNA POLYMERASE III SUBUNIT BETA, DNA POLYMERASE III SUBUNIT EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.34 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FN3
 
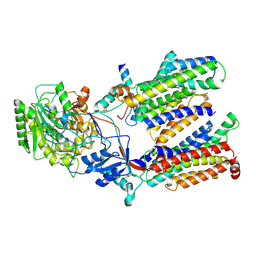 | | Cryo-EM structure of gamma secretase in class 1 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FN2
 
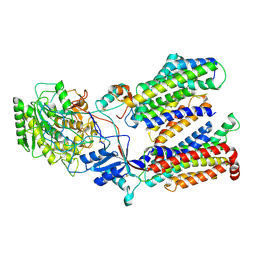 | | Cryo-EM structure of gamma secretase in complex with a drug DAPT | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
