1TNY
 
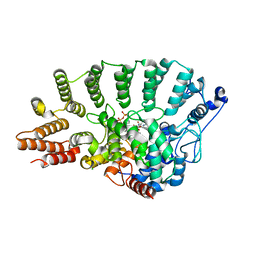 | | Rat Protein Geranylgeranyltransferase Type-I Complexed with a GGPP analog and a FREKKFFCAIL Peptide Derived from the Heterotrimeric G Protein Gamma-2 Subunit | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[METHYL-(5-GERANYL-4-METHYL-PENT-3-ENYL)-AMINO]-ETHYL-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | S Reid, T, Terry, K.L, Casey, P.J, Beese, L.S. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity.
J.Mol.Biol., 343, 2004
|
|
4WAF
 
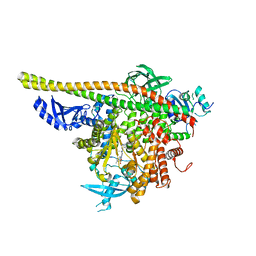 | | Crystal Structure of a novel tetrahydropyrazolo[1,5-a]pyrazine in an engineered PI3K alpha | | Descriptor: | N,N-dimethyl-4-[(6R)-6-methyl-5-(1H-pyrrolo[2,3-b]pyridin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-3-yl]benzenesulfonamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-Based Drug Design of Novel Potent and Selective Tetrahydropyrazolo[1,5-a]pyrazines as ATR Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4USO
 
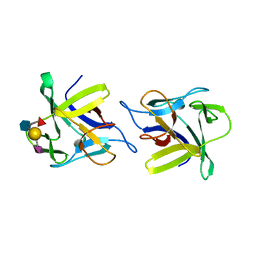 | | X-ray structure of the CCL2 lectin in complex with sialyl lewis X | | Descriptor: | CCL2 LECTIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
4W7F
 
 | |
4W85
 
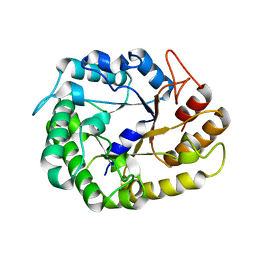 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with glucose | | Descriptor: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, beta-D-glucopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4WC5
 
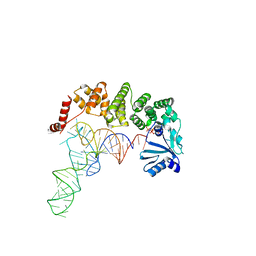 | | Structure of tRNA-processing enzyme complex 3 | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Poly A polymerase, RNA (74-MER) | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
4UZU
 
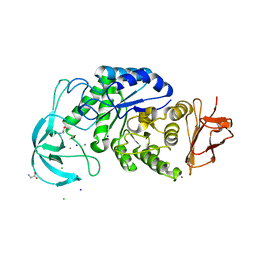 | | Three-dimensional structure of a variant `Termamyl-like' Geobacillus stearothermophilus alpha-amylase at 1.9 A resolution | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Offen, W.A, Anderson, C, Borchert, T.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of a Variant `Termamyl-Like' Geobacillus Stearothermophilus Alpha-Amylase at 1.9 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4W7H
 
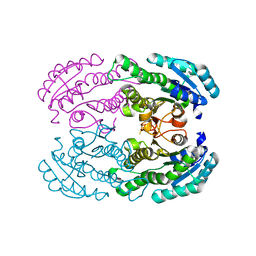 | | Crystal Structure of DEH Reductase A1-R Mutant | | Descriptor: | Carbonyl reductase | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
4W8J
 
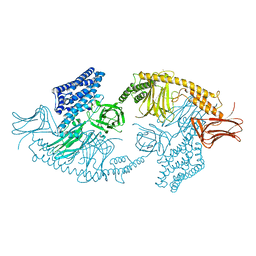 | | Structure of the full-length insecticidal protein Cry1Ac reveals intriguing details of toxin packaging into in vivo formed crystals | | Descriptor: | CALCIUM ION, POTASSIUM ION, Pesticidal crystal protein cry1Ac | | Authors: | Evdokimov, A.G, Moshiri, F, Sturman, E.J, Rydel, T.J, Zheng, M, Seale, J.W, Franklin, S. | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure of the full-length insecticidal protein Cry1Ac reveals intriguing details of toxin packaging into in vivo formed crystals.
Protein Sci., 23, 2014
|
|
4VGC
 
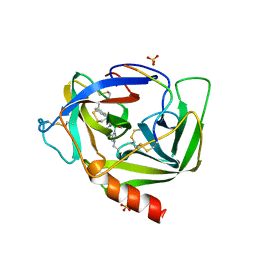 | | GAMMA-CHYMOTRYPSIN D-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | D-1-NAPHTHYL-2-ACETAMIDO-ETHANE BORONIC ACID, GAMMA CHYMOTRYPSIN, SULFATE ION | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
4V8G
 
 | | Crystal structure of RMF bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.
Science, 336, 2012
|
|
4V89
 
 | | Crystal Structure of Release Factor RF3 Trapped in the GTP State on a Rotated Conformation of the Ribosome (without viomycin) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhou, J, Lancaster, L, Trakhanov, S, Noller, H.F. | | Deposit date: | 2011-11-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome.
Rna, 18, 2012
|
|
4V90
 
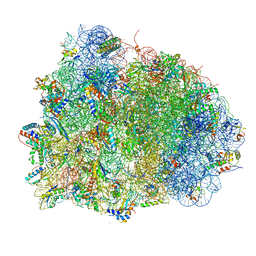 | | Thermus thermophilus Ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Chen, Y, Feng, S, Kumar, V, Ero, R, Gao, Y.G. | | Deposit date: | 2014-02-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of EF-G-ribosome complex in a pretranslocation state.
Nat. Struct. Mol. Biol., 20, 2013
|
|
4V5O
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC 40S RIBOSOMAL SUBUNIT IN COMPLEX WITH INITIATION FACTOR 1. | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S12, 40S RIBOSOMAL PROTEIN S3A, ... | | Authors: | Rabl, J, Leibundgut, M, Ataide, S.F, Haag, A, Ban, N. | | Deposit date: | 2010-11-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1.
Science, 331, 2011
|
|
4V63
 
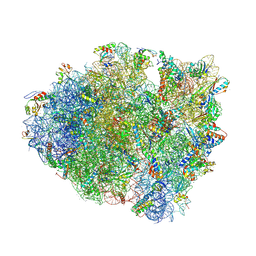 | | Structural basis for translation termination on the 70S ribosome. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Laurberg, M, Asahara, H, Korostelev, A, Zhu, J, Trakhanov, S, Noller, H.F. | | Deposit date: | 2008-05-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | Structural basis for translation termination on the 70S ribosome
Nature, 454, 2008
|
|
4WAV
 
 | | Crystal Structure of Haloquadratum walsbyi bacteriorhodopsin mutant D93N | | Descriptor: | Bacteriorhodopsin-I, RETINAL, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Wang, A.H.J, Hsu, M.F, Yang, C.S, Fu, H.Y. | | Deposit date: | 2014-09-02 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an acid-tolerant light-driven proton pump at 1.85 Angstroms resolution
To be published
|
|
4V7V
 
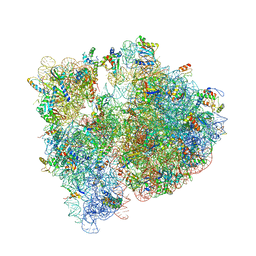 | | Crystal structure of the E. coli ribosome bound to clindamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2891 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V8K
 
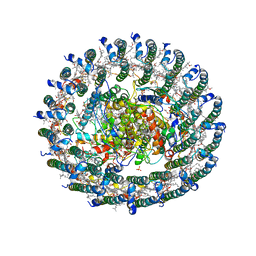 | | Crystal structure of the LH1-RC complex from Thermochromatium tepidum in P21 form | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CALCIUM ION, ... | | Authors: | Niwa, S, Takeda, K, Wang-Otomo, Z.-Y, Miki, K. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | Structure of the LH1-RC complex from Thermochromatium tepidum at 3.0 angstrom
Nature, 508, 2014
|
|
4WC8
 
 | |
4UVM
 
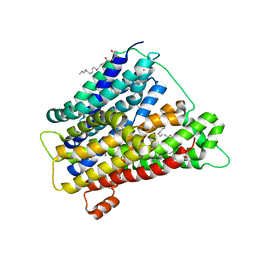 | | In meso crystal structure of the POT family transporter PepTSo | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, GLUTATHIONE UPTAKE TRANSPORTER | | Authors: | Lyons, J.A, Solcan, N, Caffrey, M, Newstead, S. | | Deposit date: | 2014-08-07 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gating Topology of the Proton-Coupled Oligopeptide Symporters.
Structure, 23, 2015
|
|
4UXU
 
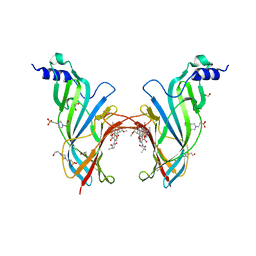 | | Crystal Structure of the Extracellular Domain of the Human Alpha9 Nicotinic Acetylcholine Receptor In Complex with Methyllycaconitine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Zouridakis, M, Giastas, P, Zarkadas, E, Tzartos, S.J. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structures of Free and Antagonist-Bound States of Human Alpha9 Nicotinic Receptor Extracellular Domain
Nat.Struct.Mol.Biol., 21, 2014
|
|
4V54
 
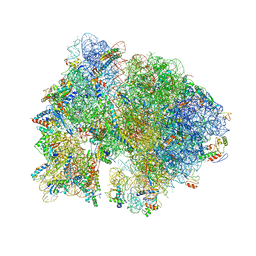 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with ribosome recycling factor (RRF). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4WBB
 
 | | Single Turnover Autophosphorylation Cycle of the PKA RIIb Holoenzyme | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Zhang, P, Knape, M.J, Ahuja, L.G, Keshwani, M.M, King, C.C, Sastri, M, Herberg, F.W, Taylor, S.S. | | Deposit date: | 2014-09-02 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Single Turnover Autophosphorylation Cycle of the PKA RII beta Holoenzyme.
Plos Biol., 13, 2015
|
|
4V24
 
 | | Sphingosine kinase 1 in complex with PF-543 | | Descriptor: | ACETATE ION, SPHINGOSINE KINASE 1, {(2R)-1-[4-({3-METHYL-5-[(PHENYLSULFONYL)METHYL]PHENOXY}METHYL)BENZYL]PYRROLIDIN-2-YL}METHANOL | | Authors: | Elkins, J.M, Wang, J, Sorrell, F, Tallant, C, Wang, D, Shrestha, L, Bountra, C, von Delft, F, Knapp, S, Edwards, A. | | Deposit date: | 2014-10-05 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Sphingosine Kinase 1 with Pf-543.
Acs Med.Chem.Lett., 5, 2014
|
|
4UQQ
 
 | | Electron density map of GluK2 desensitized state in complex with 2S,4R-4-methylglutamate | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, KAINATE 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
