7KVE
 
 | |
7KXY
 
 | |
7KVF
 
 | |
6V5T
 
 | |
6V64
 
 | | Crystal structure of human thrombin bound to ppack with tryptophans replaced by 5-F-tryptophan | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Ruben, E.A, Chen, Z, Di Cera, E. | | Deposit date: | 2019-12-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | 19F NMR reveals the conformational properties of free thrombin and its zymogen precursor prethrombin-2.
J.Biol.Chem., 295, 2020
|
|
8GBJ
 
 | | Cryo-EM structure of a human BCDX2/ssDNA complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*C)-3'), DNA repair protein RAD51 homolog 2, DNA repair protein RAD51 homolog 3, ... | | Authors: | Jia, L, Wasmuth, E.V, Ruben, E.A, Sung, P, Rawal, Y, Greene, E.C, Meir, A, Olsen, S.K. | | Deposit date: | 2023-02-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural insights into BCDX2 complex function in homologous recombination.
Nature, 619, 2023
|
|
8FY2
 
 | | E3:PROTAC:target ternary complex structure (VCB/WH244/BCL-2) | | Descriptor: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | Authors: | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Ruben, E, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
1SD0
 
 | | Structure of arginine kinase C271A mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Gattis, J.L, Ruben, E, Fenley, M.O, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2004-02-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The active site cysteine of arginine kinase: structural and functional analysis of partially active mutants
Biochemistry, 43, 2004
|
|
7TPP
 
 | |
8FAZ
 
 | | Cryo-EM structure of the human BCDX2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair protein RAD51 homolog 2, DNA repair protein RAD51 homolog 3, ... | | Authors: | Jia, L, Wasmuth, E.V, Ruben, E.A, Sung, P, Rawal, Y, Greene, E.C, Meir, A, Olsen, S.K. | | Deposit date: | 2022-11-29 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural insights into BCDX2 complex function in homologous recombination.
Nature, 619, 2023
|
|
6V09
 
 | | Crystal structure of human recombinant Beta-2 glycoprotein I short tag (ST-B2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
3NBR
 
 | | Crystal Structure of Ketosteroid Isomerase D38NP39GD99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Ruben, E, Sunden, F, Herschlag, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D38NP39GD99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound
To be Published
|
|
3NHX
 
 | | Crystal Structure of Ketosteroid Isomerase D99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Ruben, E, Sunden, F, Herschlag, D. | | Deposit date: | 2010-06-14 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound
To be Published
|
|
6V06
 
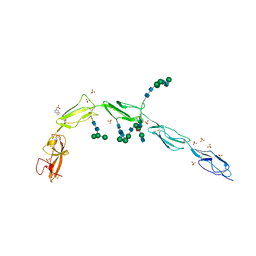 | | Crystal structure of Beta-2 glycoprotein I purified from plasma (pB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-glycoprotein 1, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
6V08
 
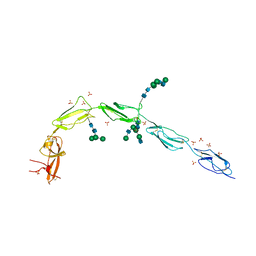 | | Crystal structure of human recombinant Beta-2 glycoprotein I (hrB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
8TWK
 
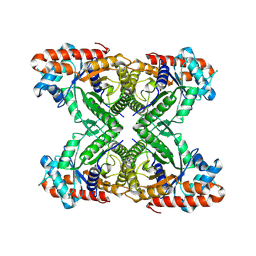 | |
8TWL
 
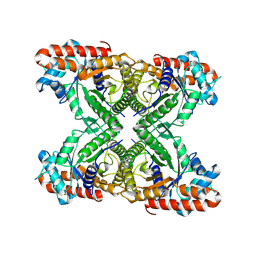 | |
8TUE
 
 | |
8TWM
 
 | |
8SE9
 
 | | Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 2) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | Authors: | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | Deposit date: | 2023-04-08 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
8SV8
 
 | | Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex from a composite map | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | Authors: | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
8SEA
 
 | | Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 1) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | Authors: | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | Deposit date: | 2023-04-08 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
8SEB
 
 | | Cryo-EM structure of a single loaded human UBA7-UBE2L6-ISG15 adenylate complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | Authors: | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | Deposit date: | 2023-04-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
7SR9
 
 | | Human alpha-thrombin with 180- and 220- loops replaced with homologous loops from protein C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Di Cera, E, Ruben, E.A, Chen, Z. | | Deposit date: | 2021-11-08 | | Release date: | 2021-12-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The active site region plays a critical role in Na + binding to thrombin.
J.Biol.Chem., 298, 2022
|
|
