6WJ5
 
 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
6VJA
 
 | | Structure of CD20 in complex with rituximab Fab | | Descriptor: | B-lymphocyte antigen CD20, CHOLESTEROL HEMISUCCINATE, Rituximab Fab heavy chain, ... | | Authors: | Rohou, A, Croll, T.I. | | Deposit date: | 2020-01-15 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of CD20 in complex with the therapeutic monoclonal antibody rituximab.
Science, 367, 2020
|
|
6X2J
 
 | | Structure of human TRPA1 in complex with agonist GNE551 | | Descriptor: | 5-amino-1-[(4-bromo-2-fluorophenyl)methyl]-N-(2,5-dimethoxyphenyl)-1H-1,2,3-triazole-4-carboxamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Chen, H. | | Deposit date: | 2020-05-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A Non-covalent Ligand Reveals Biased Agonism of the TRPA1 Ion Channel.
Neuron, 109, 2021
|
|
7JUP
 
 | | Structure of human TRPA1 in complex with antagonist compound 21 | | Descriptor: | 1-({3-[(3R,5R)-5-(4-fluorophenyl)oxolan-3-yl]-1,2,4-oxadiazol-5-yl}methyl)-7-methyl-1,7-dihydro-6H-purin-6-one, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L. | | Deposit date: | 2020-08-20 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Tetrahydrofuran-Based Transient Receptor Potential Ankyrin 1 (TRPA1) Antagonists: Ligand-Based Discovery, Activity in a Rodent Asthma Model, and Mechanism-of-Action via Cryogenic Electron Microscopy.
J.Med.Chem., 64, 2021
|
|
5SV9
 
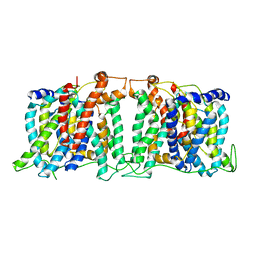 | | Structure of the SLC4 transporter Bor1p in an inward-facing conformation | | Descriptor: | Bor1p boron transporter | | Authors: | Coudray, N, Seyler, S, Lasala, R, Zhang, Z, Clark, K.M, Dumont, M.E, Rohou, A, Beckstein, O, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | Deposit date: | 2016-08-05 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structure of the SLC4 transporter Bor1p in an inward-facing conformation.
Protein Sci., 26, 2017
|
|
5TJ5
 
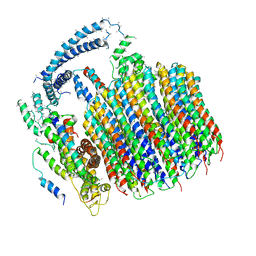 | | Atomic model for the membrane-embedded motor of a eukaryotic V-ATPase | | Descriptor: | V-type proton ATPase subunit a, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | Authors: | Mazhab-Jafari, M.T, Rohou, A, Schmidt, C, Bueler, S.A, Benlekbir, S, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2016-10-03 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase.
Nature, 539, 2016
|
|
2VY9
 
 | | Molecular architecture of the stressosome, a signal integration and transduction hub | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST | | Authors: | Marles-Wright, J, Grant, T, Delumeau, O, van Duinen, G, Firbank, S.J, Lewis, P.J, Murray, J.W, Newman, J.A, Quin, M.B, Race, P.R, Rohou, A, Tichelaar, W, van Heel, M, Lewis, R.J. | | Deposit date: | 2008-07-21 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Architecture of the "Stressosome," a Signal Integration and Transduction Hub
Science, 322, 2008
|
|
8F0S
 
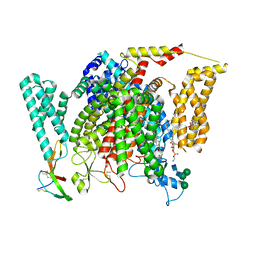 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-9296 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0Q
 
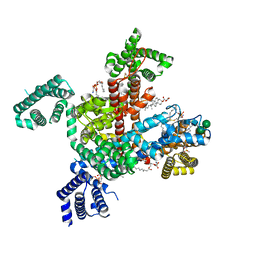 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acylsulfonamide inhibitor GDC-0310 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-cyclopropyl-4-({1-[(1S)-1-(3,5-dichlorophenyl)ethyl]piperidin-4-yl}methoxy)-2-fluoro-N-(methanesulfonyl)benzamide, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0P
 
 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-1305 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0R
 
 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the arylsulfonamide inhibitor GNE-3565 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-4-({(1S,2S,4S)-2-(dimethylamino)-4-[3-(trifluoromethyl)phenyl]cyclohexyl}amino)-2-fluoro-N-(pyrimidin-4-yl)benzene-1-sulfonamide, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
5FIK
 
 | | Bovine mitochondrial ATP synthase state 3a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
5FIJ
 
 | | Bovine mitochondrial ATP synthase state 2c | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
5FIL
 
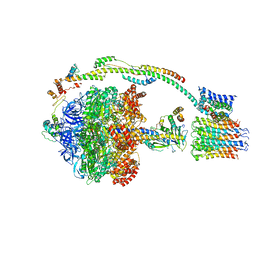 | | Bovine mitochondrial ATP synthase state 3b | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
7LBE
 
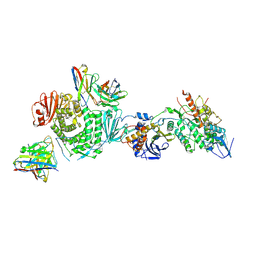 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBG
 
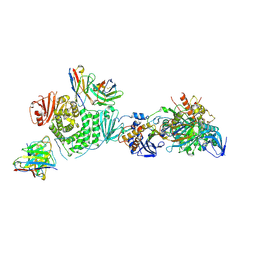 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Transforming growth factor beta receptor type 3 and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBF
 
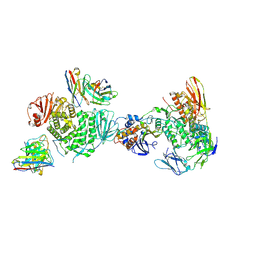 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Platelet-derived growth factor receptor alpha and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
6N4Q
 
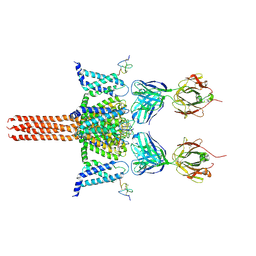 | | CryoEM structure of Nav1.7 VSD2 (actived state) in complex with the gating modifier toxin ProTx2 | | Descriptor: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | Authors: | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
6N2P
 
 | | Helical assembly of the CARD9 CARD | | Descriptor: | Caspase recruitment domain-containing protein 9 | | Authors: | Holliday, M.J, Rohou, A, Arthur, C.P, Dueber, E.C, Fairbrother, W.J. | | Deposit date: | 2018-11-13 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of autoinhibited and polymerized forms of CARD9 reveal mechanisms of CARD9 and CARD11 activation.
Nat Commun, 10, 2019
|
|
6N4R
 
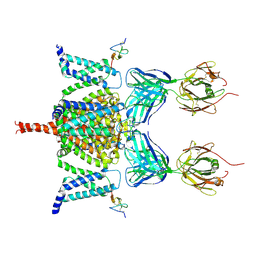 | | CryoEM structure of Nav1.7 VSD2 (deactived state) in complex with the gating modifier toxin ProTx2 | | Descriptor: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | Authors: | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
7T4Q
 
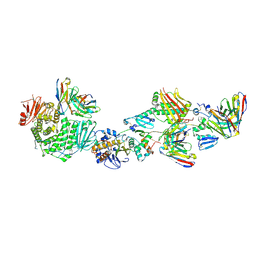 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with neutralizing fabs 2C12, 7I13 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4R
 
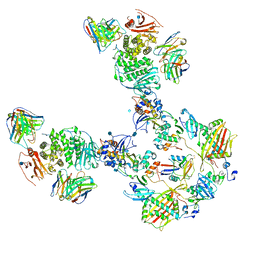 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with THBD and neutralizing fabs MSL-109 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4S
 
 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with NRP2 and neutralizing fabs 8I21 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
5ARH
 
 | | Bovine mitochondrial ATP synthase state 2a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
5ARA
 
 | | Bovine mitochondrial ATP synthase state 1a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
