2X6G
 
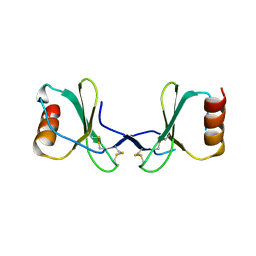 | |
2X6L
 
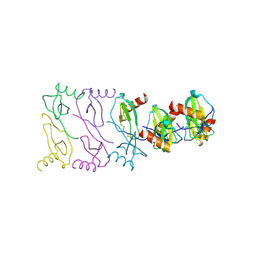 | |
7RK6
 
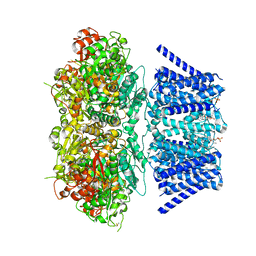 | | Aplysia Slo1 with Barium | | 分子名称: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, BARIUM ION, BK channel, ... | | 著者 | Zhu, J, Srivastava, S, Cachau, R, Holmgren, M. | | 登録日 | 2021-07-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-04-05 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | CryoEM structure of Aplysia Slo1 with 0 mM Ba2+ at 2.91 A
To Be Published
|
|
7RJT
 
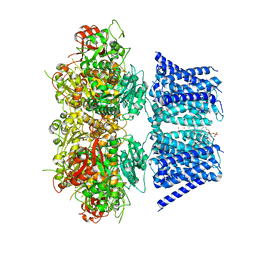 | | Aplysia Slo1 with Barium | | 分子名称: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, BARIUM ION, BK channel, ... | | 著者 | Zhu, J, Srivastava, S, Cachau, R, Holmgren, M. | | 登録日 | 2021-07-21 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | CryoEM structure of Aplysia Slo1 with 10 mM Ba2+ at 2.93 A
To Be Published
|
|
3KBX
 
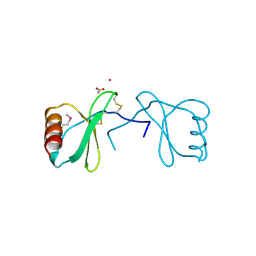 | |
2X69
 
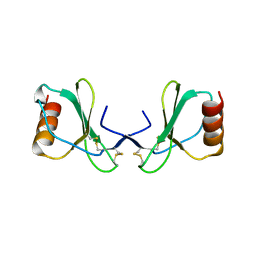 | |
7MQS
 
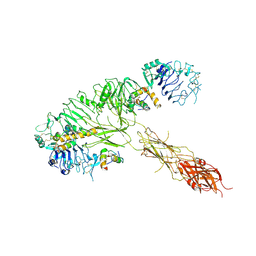 | | The insulin receptor ectodomain in complex with three venom hybrid insulin molecules - asymmetric conformation | | 分子名称: | Insulin A chain, Insulin B chain, Isoform Short of Insulin receptor | | 著者 | Blakely, A.D, Xiong, X, Kim, J.H, Menting, J, Schafer, I.B, Schubert, H.L, Agrawal, R, Gutmann, T, Delaine, C, Zhang, Y, Artik, G.O, Merriman, A, Eckert, D, Lawrence, M.C, Coskun, U, Fisher, S.J, Forbes, B.E, Safavi-Hemami, H, Hill, C.P, Chou, D.H.C. | | 登録日 | 2021-05-06 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Symmetric and asymmetric receptor conformation continuum induced by a new insulin.
Nat.Chem.Biol., 18, 2022
|
|
1MOX
 
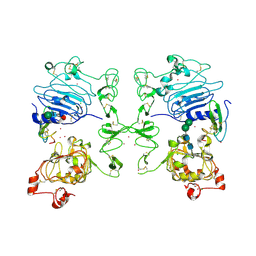 | | Crystal Structure of Human Epidermal Growth Factor Receptor (residues 1-501) in complex with TGF-alpha | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Garrett, T.P.J, McKern, N.M, Lou, M, Elleman, T.C, Adams, T.E, Lovrecz, G.O, Zhu, H.-J, Walker, F, Frenkel, M.J, Hoyne, P.A, Jorissen, R.N, Nice, E.C, Burgess, A.W, Ward, C.W. | | 登録日 | 2002-09-10 | | 公開日 | 2003-09-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of a Truncated Epidermal Growth Factor Receptor Extracellular Domain Bound to Transforming Growth Factor alpha
Cell(Cambridge,Mass.), 110, 2002
|
|
4V4B
 
 | | Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map. | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11, ... | | 著者 | Spahn, C.M, Gomez-Lorenzo, M.G, Grassucci, R.A, Jorgensen, R, Andersen, G.R, Beckmann, R, Penczek, P.A, Ballesta, J.P.G, Frank, J. | | 登録日 | 2004-01-06 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (11.7 Å) | | 主引用文献 | Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation.
Embo J., 23, 2004
|
|
1ML5
 
 | | Structure of the E. coli ribosomal termination complex with release factor 2 | | 分子名称: | 30S 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | 著者 | Klaholz, B.P, Pape, T, Zavialov, A.V, Myasnikov, A.G, Orlova, E.V, Vestergaard, B, Ehrenberg, M, van Heel, M. | | 登録日 | 2002-08-30 | | 公開日 | 2003-01-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (14 Å) | | 主引用文献 | Structure of the Escherichia coli ribosomal termination complex with release factor 2
Nature, 421, 2003
|
|
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | 分子名称: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | 著者 | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | 登録日 | 2017-03-23 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
4UN1
 
 | | Sirohaem decarboxylase AhbA/B - an enzyme with structural homology to the Lrp/AsnC transcription factor family that is part of the alternative haem biosynthesis pathway. | | 分子名称: | 12,18-DIDECARBOXY-SIROHEME, PUTATIVE TRANSCRIPTIONAL REGULATOR, ASNC FAMILY | | 著者 | Palmer, D.J, Brown, D.G, Warren, M.J, Pickersgill, R.W. | | 登録日 | 2014-05-23 | | 公開日 | 2014-06-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | The Structure, Function and Properties of Sirohaem Decarboxylase - an Enzyme with Structural Homology to a Transcription Factor Family that is Part of the Alternative Haem Biosynthesis Pathway.
Mol.Microbiol., 93, 2014
|
|
9EUU
 
 | | Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo | | 分子名称: | Alpha-synuclein | | 著者 | Burger, D, Kashyrina, M, Lewis, A, De Nuccio, F, Mohammed, I, de La Seigliere, H, van den Heuvel, L, Feuillie, C, Verchere, J, Berbon, M, Arotcarena, M, Retailleau, A, Bezard, E, Laferriere, F, Loquet, A, Bousset, L, Baron, T, Lofrumento, D.D, De Giorgi, F, Stahlberg, H, Ichas, F. | | 登録日 | 2024-03-28 | | 公開日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (1.93 Å) | | 主引用文献 | Multiple System Atrophy: Insights from aSyn Fibril Structure
To Be Published
|
|
4XSS
 
 | | Insulin-like growth factor I in complex with site 1 of a hybrid insulin receptor / Type 1 insulin-like growth factor receptor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin receptor, ... | | 著者 | Lawrence, C, Kong, G.K.-W, Menting, J.G, Lawrence, M.C. | | 登録日 | 2015-01-22 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Congruency of Ligand Binding to the Insulin and Insulin/Type 1 Insulin-like Growth Factor Hybrid Receptors.
Structure, 23, 2015
|
|
6QI4
 
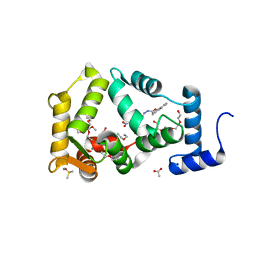 | | NCS-1 bound to a ligand | | 分子名称: | 2-(1~{H}-indol-3-yl)-~{N}-[(~{E})-(4-nitro-3-oxidanyl-phenyl)methylideneamino]ethanamide, ACETATE ION, CALCIUM ION, ... | | 著者 | Sanchez-Barrena, M.J, Blanco-Gabella, P. | | 登録日 | 2019-01-17 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Insights into real-time chemical processes in a calcium sensor protein-directed dynamic library.
Nat Commun, 10, 2019
|
|
4XST
 
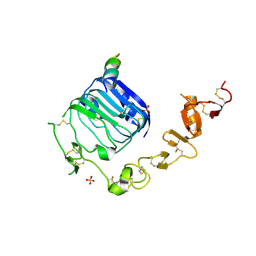 | | Structure of the endoglycosidase-H treated L1-CR domains of the human insulin receptor in complex with residues 697-719 of the human insulin receptor (A-isoform) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin receptor, SULFATE ION, ... | | 著者 | Menting, J.G, Lawrence, C.F, Kong, G.K.-W, Lawrence, M.C. | | 登録日 | 2015-01-22 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Congruency of Ligand Binding to the Insulin and Insulin/Type 1 Insulin-like Growth Factor Hybrid Receptors.
Structure, 23, 2015
|
|
8ALH
 
 | |
6NUL
 
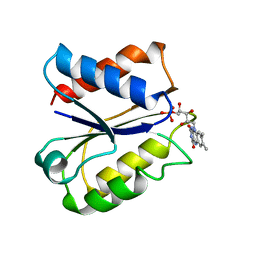 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: N137A REDUCED (150K) | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | 著者 | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | 登録日 | 1997-01-09 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
8ALM
 
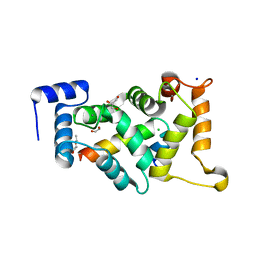 | |
8AHY
 
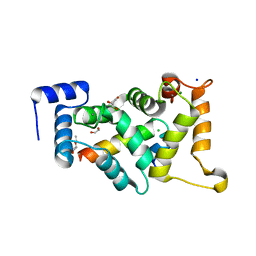 | |
6NE0
 
 | | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | | 分子名称: | CRISPR RNA (60-MER), CRISPR target DNA (44-MER), CRISPR-associated endonuclease Cas6/Csy4, ... | | 著者 | Chowdhury, S, Rollins, M.F, Carter, J, Golden, S.M, Miettinen, H.M, Santiago-Frangos, A, Faith, D, Lawrence, M.C, Wiedenheft, B, Lander, G.C. | | 登録日 | 2018-12-15 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Mol. Cell, 74, 2019
|
|
6EPA
 
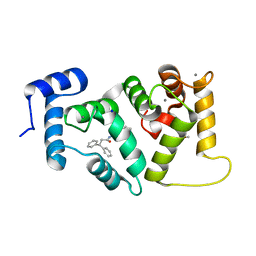 | | Structure of dNCS-1 bound to the NCS-1/Ric8a protein/protein interaction regulator IGS-1.76 | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, FI18190p1, ... | | 著者 | Sanchez-Barrena, M.J, Daniel, M, Infantes, L. | | 登録日 | 2017-10-11 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Deciphering the Inhibition of the Neuronal Calcium Sensor 1 and the Guanine Exchange Factor Ric8a with a Small Phenothiazine Molecule for the Rational Generation of Therapeutic Synapse Function Regulators.
J. Med. Chem., 61, 2018
|
|
5VY8
 
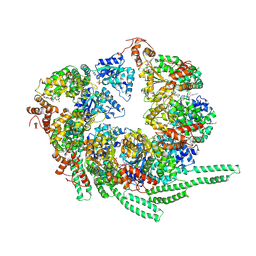 | | S. cerevisiae Hsp104-ADP complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 104 | | 著者 | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | 登録日 | 2017-05-24 | | 公開日 | 2017-07-05 | | 最終更新日 | 2018-08-15 | | 実験手法 | ELECTRON MICROSCOPY (5.6 Å) | | 主引用文献 | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
7TPG
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state | | 分子名称: | Fab Heavy (H) Chain, Fab Light (L) Chain, GERANYL DIPHOSPHATE, ... | | 著者 | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | 登録日 | 2022-01-25 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-04-27 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
7TPJ
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state | | 分子名称: | Fab Heavy (H) Chain, Fab Light (L) Chain, Putative cell surface polysaccharide polymerase/ligase | | 著者 | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | 登録日 | 2022-01-25 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-04-27 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
