1EJ9
 
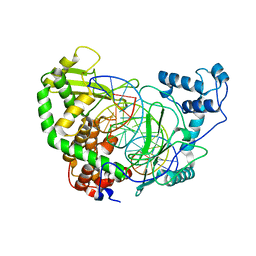 | | CRYSTAL STRUCTURE OF HUMAN TOPOISOMERASE I DNA COMPLEX | | Descriptor: | DNA (5'-D(*C*AP*AP*AP*AP*AP*GP*AP*CP*TP*CP*AP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3'), DNA (5'-D(*C*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*GP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), DNA TOPOISOMERASE I | | Authors: | Redinbo, M.R, Champoux, J.J, Hol, W.G. | | Deposit date: | 2000-03-01 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel insights into catalytic mechanism from a crystal structure of human topoisomerase I in complex with DNA.
Biochemistry, 39, 2000
|
|
2PLT
 
 | |
1A35
 
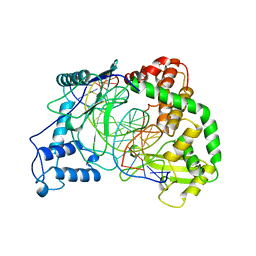 | | HUMAN TOPOISOMERASE I/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*AP*(BRU)P*(BRU)P*TP*TP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*+UP*+UP*+UP*+UP*CP*+UP*AP*AP*GP*TP*CP*TP*TP*TP*+ UP*T)-3'), PROTEIN (DNA TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G. | | Deposit date: | 1998-01-29 | | Release date: | 1998-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
1A31
 
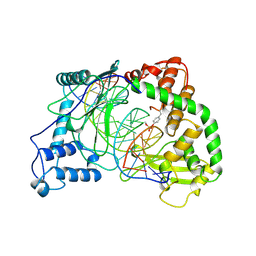 | | HUMAN RECONSTITUTED DNA TOPOISOMERASE I IN COVALENT COMPLEX WITH A 22 BASE PAIR DNA DUPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*5IUP*5IU*TP*GP*AP*AP*AP*AP*AP*5IUP*5IUP*5IUP*5IUP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*5IUP*5IUP*5IUP*5IUP*CP*AP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), PROTEIN (TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 1998-01-27 | | Release date: | 1998-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
3L6T
 
 | |
3L57
 
 | |
3HX6
 
 | |
7U4C
 
 | | Borrelia burgdorferi HtpG N-terminal domain (1-228) in complex with BX-2819 | | Descriptor: | Chaperone protein HtpG, ethyl (4-{3-[2,4-dihydroxy-5-(1-methylethyl)phenyl]-5-sulfanyl-4H-1,2,4-triazol-4-yl}benzyl)carbamate | | Authors: | Kowalewski, M.E, Lietzan, A, Haystead, T, Redinbo, M.R. | | Deposit date: | 2022-02-28 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Targeting Borrelia burgdorferi HtpG with a berserker molecule, a strategy for anti-microbial development.
Cell Chem Biol, 2023
|
|
3K46
 
 | |
6U7J
 
 | | Uncultured Clostridium sp. Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CALCIUM ION | | Authors: | Ervin, S.M, Redinbo, M.R. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gut microbial beta-glucuronidases reactivate estrogens as components of the estrobolome that reactivate estrogens.
J.Biol.Chem., 294, 2019
|
|
2A66
 
 | | Human Liver Receptor Homologue DNA-Binding Domain (hLRH-1 DBD) in Complex with dsDNA from the hCYP7A1 Promoter | | Descriptor: | 5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3', 5'-D(*GP*TP*TP*CP*AP*AP*GP*GP*CP*CP*AP*G)-3', ACETATE ION, ... | | Authors: | Solomon, I.H, Hager, J.M, Safi, R, McDonnell, D.P, Redinbo, M.R, Ortlund, E.A. | | Deposit date: | 2005-07-01 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human LRH-1 DBD-DNA Complex Reveals Ftz-F1 Domain Positioning is Required for Receptor Activity.
J.Mol.Biol., 354, 2005
|
|
8DHL
 
 | | Tannerella forsythia beta-glucuronidase (L2) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 2, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-27 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8DHW
 
 | | Treponema lecithinolyticum beta-glucuronidase in complex with a UNC4917-glucuronide conjugate | | Descriptor: | 4-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-2,7-bis(methylamino)pyrido[3',2':4,5]thieno[3,2-d]pyrimidine, Glycosyl hydrolase family 2, TIM barrel domain protein, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8DHV
 
 | | Treponema lecithinolyticum beta-glucuronidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Glycosyl hydrolase family 2, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8DHE
 
 | |
6P2B
 
 | | Tethered PXR-LBD/SRC-1p bound to Garcinoic Acid | | Descriptor: | (2Z,6E,10E)-13-[(2R)-6-hydroxy-2,8-dimethyl-3,4-dihydro-2H-1-benzopyran-2-yl]-2,6,10-trimethyltrideca-2,6,10-trienoic acid, DIMETHYL SULFOXIDE, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2019-05-21 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Garcinoic Acid Is a Natural and Selective Agonist of Pregnane X Receptor.
J.Med.Chem., 63, 2020
|
|
8UGT
 
 | | E. eligens beta-glucuronidase bound to UNC10206581-G | | Descriptor: | 8-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5-(methylamino)-1,2,3,4-tetrahydro[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinoline, Beta-glucuronidase, GLYCEROL, ... | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-10-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advanced piperazine-containing inhibitors target microbial beta-glucuronidases linked to gut toxicity.
Rsc Chem Biol, 5, 2024
|
|
8UMG
 
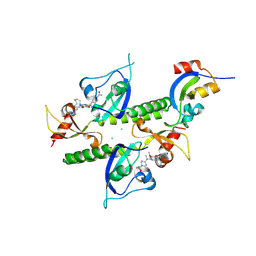 | | Chromodomains of human CHD1 complexed with UNC10142 | | Descriptor: | 1-{4-[{2-(azonan-1-yl)-6-methoxy-7-[3-(piperidin-1-yl)propoxy]quinazolin-4-yl}(methyl)amino]piperidin-1-yl}ethan-1-one, CHLORIDE ION, Chromodomain-helicase-DNA-binding protein 1 | | Authors: | Graboski, A.L, Redinbo, M.R. | | Deposit date: | 2023-10-17 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Optimization of a quinazoline-based scaffold against human CHD1
To Be Published
|
|
8W1Q
 
 | | Aerobic crystal structure of iron-bound FlcD from Pseudomonas aeruginosa | | Descriptor: | FE (II) ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Walker, M.E, Grove, T.L, Li, B, Redinbo, M.R. | | Deposit date: | 2024-02-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Basis for Methine Excision by a Heme Oxygenase-like Enzyme.
Acs Cent.Sci., 10, 2024
|
|
5CZK
 
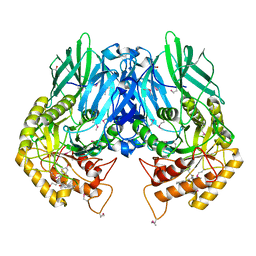 | | Structure of E. coli beta-glucuronidase bound with a novel, potent inhibitor 1-((6,8-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl)-1-(2-hydroxyethyl)-3-(4-hydroxyphenyl)thiourea | | Descriptor: | 1-[(6,8-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl]-1-(2-hydroxyethyl)-3-(4-hydroxyphenyl)thiourea, Beta-glucuronidase | | Authors: | Roberts, A.R, Wallace, B.R, Redinbo, M.R. | | Deposit date: | 2015-07-31 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure and Inhibition of Microbiome beta-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity.
Chem.Biol., 22, 2015
|
|
8SBG
 
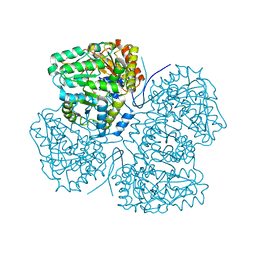 | |
8SIJ
 
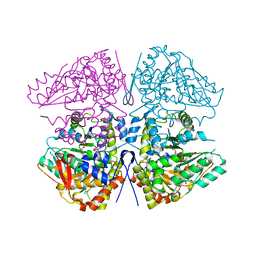 | | Crystal structure of F. varium tryptophanase | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Tryptophanase 1, ... | | Authors: | Graboski, A.L, Redinbo, M.R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism-based inhibition of gut microbial tryptophanases reduces serum indoxyl sulfate.
Cell Chem Biol, 30, 2023
|
|
8SL7
 
 | | Butyricicoccus sp. BIOML-A1 tryptophanase complex with (3S) ALG-05 | | Descriptor: | (E)-3-[(3S)-3-chloro-2-oxo-2,3-dihydro-1H-indol-3-yl]-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-alanine, Tryptophanase | | Authors: | Graboski, A.L, Redinbo, M.R. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Mechanism-based inhibition of gut microbial tryptophanases reduces serum indoxyl sulfate.
Cell Chem Biol, 30, 2023
|
|
9B9M
 
 | | Crystal structure of iron-bound FlcD from Pseudomonas aeruginosa | | Descriptor: | FE (II) ION, Pyrroloquinoline quinone (Coenzyme PQQ) biosynthesis protein C | | Authors: | Walker, M.E, Grove, T.L, Li, B, Redinbo, M.R. | | Deposit date: | 2024-04-02 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for Methine Excision by a Heme Oxygenase-like Enzyme.
Acs Cent.Sci., 10, 2024
|
|
9B9O
 
 | | Crystal structure of FlcD from Pseudomonas aeruginosa bond to iron(II) and substrate | | Descriptor: | (2R)-2-{[(2Z)-2-(hydroxyimino)ethyl]sulfanyl}butanedioic acid, FE (II) ION, Pyrroloquinoline quinone (Coenzyme PQQ) biosynthesis protein C | | Authors: | Walker, M.E, Grove, T.L, Li, B, Redinbo, M.R. | | Deposit date: | 2024-04-02 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis for Methine Excision by a Heme Oxygenase-like Enzyme.
Acs Cent.Sci., 10, 2024
|
|
