1XF9
 
 | | Structure of NBD1 from murine CFTR- F508S mutant | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Thibodeau, P.H, Brautigam, C.A, Machius, M, Thomas, P.J. | | Deposit date: | 2004-09-14 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Side chain and backbone contributions of Phe508 to CFTR folding.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2YN3
 
 | | Structural insight into the giant calcium-binding adhesin SiiE: implications for the adhesion of Salmonella enterica to polarized epithelial cells | | Descriptor: | CALCIUM ION, IODIDE ION, PHOSPHATE ION, ... | | Authors: | Griessl, M.H, Schmid, B, Kassler, K, Braunsmann, C, Ritter, R, Barlag, B, Sturm, K.U, Danzer, C, Wagner, C, Schaeffer, T.E, Sticht, H, Hensel, M, Muller, Y.A. | | Deposit date: | 2012-10-11 | | Release date: | 2013-04-03 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Insight Into the Giant Ca(2+)-Binding Adhesin Siie: Implications for the Adhesion of Salmonella Enterica to Polarized Epithelial Cells.
Structure, 21, 2013
|
|
6VGC
 
 | | Crystal Structures of FLAP bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, 5-lipoxygenase-activating protein, CALCIUM ION, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6VL4
 
 | | Crystal Structure of mPGES-1 bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, Prostaglandin E synthase, TETRAETHYLENE GLYCOL, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-22 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
1XFA
 
 | | Structure of NBD1 from murine CFTR- F508R mutant | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Thibodeau, P.H, Brautigam, C.A, Machius, M, Thomas, P.J. | | Deposit date: | 2004-09-14 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Side chain and backbone contributions of Phe508 to CFTR folding.
Nat.Struct.Mol.Biol., 12, 2005
|
|
3G9A
 
 | | Green fluorescent protein bound to minimizer nanobody | | Descriptor: | Green fluorescent protein, Minimizer | | Authors: | Kirchhofer, A, Helma, J, Schmidthals, K, Frauer, C, Cui, S, Karcher, A, Pellis, M, Muyldermans, S, Delucci, C.C, Cardoso, M.C, Leonhardt, H, Hopfner, K.-P, Rothbauer, U. | | Deposit date: | 2009-02-13 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.614 Å) | | Cite: | Modulation of protein properties in living cells using nanobodies
Nat.Struct.Mol.Biol., 17, 2010
|
|
4M3F
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | HTH-type transcriptional regulator EthR, N-(3-methylbutyl)-4-(2-methyl-1,3-thiazol-4-yl)benzenesulfonamide | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
2YN5
 
 | | Structural insight into the giant calcium-binding adhesin SiiE: implications for the adhesion of Salmonella enterica to polarized epithelial cells | | Descriptor: | CALCIUM ION, PUTATIVE INNER MEMBRANE PROTEIN | | Authors: | Griessl, M.H, Schmid, B, Kassler, K, Braunsmann, C, Ritter, R, Barlag, B, Sturm, K.U, Danzer, C, Wagner, C, Schaeffer, T.E, Sticht, H, Hensel, M, Muller, Y.A. | | Deposit date: | 2012-10-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insight Into the Giant Ca(2+)-Binding Adhesin Siie: Implications for the Adhesion of Salmonella Enterica to Polarized Epithelial Cells.
Structure, 21, 2013
|
|
4M3D
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-{3-[(phenylsulfonyl)amino]prop-1-yn-1-yl}-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
4M3E
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-{[(propylsulfonyl)amino]methyl}-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
2UXN
 
 | | Structural Basis of Histone Demethylation by LSD1 Revealed by Suicide Inactivation | | Descriptor: | CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Szewczuk, L.M, Gocke, C.B, Brautigam, C.A, Tomchick, D.R, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Histone Demethylation by Lsd1 Revealed by Suicide Inactivation.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4M3G
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-methyl-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzenesulfonamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
4M8N
 
 | | Crystal Structure of PlexinC1/Rap1B Complex | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pascoe, H.G, Wang, Y, Brautigam, C.A, He, H, Zhang, X. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin.
Elife, 2, 2013
|
|
3V5Y
 
 | | Structure of FBXL5 hemerythrin domain, P2(1) cell | | Descriptor: | F-box/LRR-repeat protein 5, MU-OXO-DIIRON | | Authors: | Tomchick, D.R, Bruick, R.K, Thompson, J.W, Brautigam, C.A. | | Deposit date: | 2011-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
J.Biol.Chem., 287, 2012
|
|
2QJO
 
 | | crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase (NadM) complexed with ADPRP and NAD from Synechocystis sp. | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Bifunctional NMN adenylyltransferase/Nudix hydrolase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Raffaelli, N, Magni, G, Grishin, N.V, Osterman, A, Zhang, H. | | Deposit date: | 2007-07-08 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
2II5
 
 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), Isobutyryl-Coenzyme A-bound form | | Descriptor: | ACETATE ION, CHLORIDE ION, ISOBUTYRYL-COENZYME A, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
2IHW
 
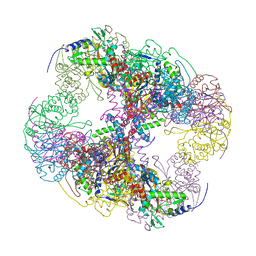 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), apo form | | Descriptor: | ACETATE ION, CHLORIDE ION, Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
1II6
 
 | | Crystal Structure of the Mitotic Kinesin Eg5 in Complex with Mg-ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-RELATED MOTOR PROTEIN Eg5, MAGNESIUM ION, ... | | Authors: | Turner, J, Anderson, R, Guo, J, Beraud, C, Sakowicz, R, Fletterick, R. | | Deposit date: | 2001-04-20 | | Release date: | 2001-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker.
J.Biol.Chem., 276, 2001
|
|
3CPU
 
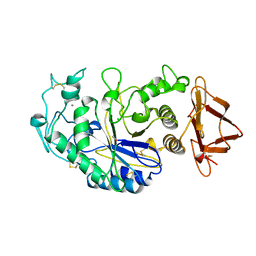 | | SUBSITE MAPPING OF THE ACTIVE SITE OF HUMAN PANCREATIC ALPHA-AMYLASE USING SUBSTRATES, THE PHARMACOLOGICAL INHIBITOR ACARBOSE, AND AN ACTIVE SITE VARIANT | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Brayer, G.D, Sidhu, G, Maurus, R, Rydberg, E.H, Braun, C, Wang, Y, Nguyen, N.T, Overall, C.M, Withers, S.G. | | Deposit date: | 1999-06-08 | | Release date: | 2001-06-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subsite mapping of the human pancreatic alpha-amylase active site through structural, kinetic, and mutagenesis techniques.
Biochemistry, 39, 2000
|
|
1XS5
 
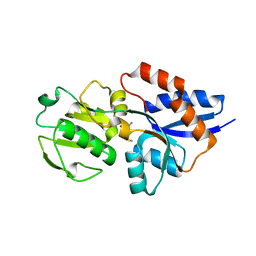 | | The Crystal Structure of Lipoprotein Tp32 from Treponema pallidum | | Descriptor: | METHIONINE, Membrane lipoprotein TpN32 | | Authors: | Deka, R.K, Neil, L, Hagman, K.E, Machius, M, Tomchick, D.R, Brautigam, C.A, Norgard, M.V. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural evidence that the 32-kilodalton lipoprotein (Tp32) of Treponema pallidum is an L-methionine-binding protein
J.Biol.Chem., 279, 2004
|
|
4M3B
 
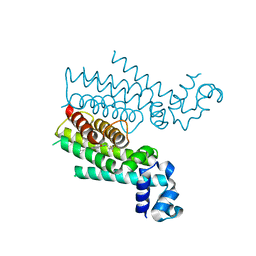 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-methyl-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
3V5Z
 
 | | Structure of FBXL5 hemerythrin domain, C2 cell, grown anaerobically | | Descriptor: | F-box/LRR-repeat protein 5, MU-OXO-DIIRON | | Authors: | Tomchick, D.R, Bruick, R.K, Thompson, J.W, Brautigam, C.A. | | Deposit date: | 2011-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1847 Å) | | Cite: | Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
J.Biol.Chem., 287, 2012
|
|
3V5X
 
 | | Structure of FBXL5 hemerythrin domain, C2 cell | | Descriptor: | F-box/LRR-repeat protein 5, MU-OXO-DIIRON | | Authors: | Tomchick, D.R, Bruick, R.K, Thompson, J.W, Brautigam, C.A. | | Deposit date: | 2011-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
J.Biol.Chem., 287, 2012
|
|
2CMN
 
 | | A Proximal Arginine Residue in the Switching Mechanism of the FixL Oxygen Sensor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL | | Authors: | Gilles-Gonzalez, M.-A, Caceres, A.I, Silva Sousa, E.H, Tomchick, D.R, Brautigam, C.A, Gonzalez, C, Machius, M. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Proximal Arginine R206 Participates in Switching of the Bradyrhizobium Japonicum Fixl Oxygen Sensor
J.Mol.Biol., 360, 2006
|
|
4FFB
 
 | | A TOG:alpha/beta-tubulin Complex Structure Reveals Conformation-Based Mechanisms For a Microtubule Polymerase | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein STU2, ... | | Authors: | Ayaz, P, Ye, X, Huddleston, P, Brautigam, C.A, Rice, L.M. | | Deposit date: | 2012-05-31 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | A TOG: alpha beta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase.
Science, 337, 2012
|
|
