8FIR
 
 | | Crystal structure of TpPta, a phosphotransacetylase from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Phosphate acetyltransferase | | Authors: | Brautigam, C.A, Norgard, M.V, Deka, R.K. | | Deposit date: | 2022-12-16 | | Release date: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biophysical and biochemical studies support TP0094 as a phosphotransacetylase in an acetogenic energy-conservation pathway in Treponema pallidum.
Plos One, 18, 2023
|
|
4U3Q
 
 | |
5JX2
 
 | | Crystal structure of MglB-2 (Tp0684) from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2016-05-12 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.0504 Å) | | Cite: | The Tp0684 (MglB-2) Lipoprotein of Treponema pallidum: A Glucose-Binding Protein with Divergent Topology.
Plos One, 11, 2016
|
|
6DET
 
 | |
1QSL
 
 | |
7MGT
 
 | | Ftp from Treponema pallidum bound to an ADP-like inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-chloroadenosine 5'-(trihydrogen diphosphate), FAD:protein FMN transferase, ... | | Authors: | Brautigam, C.A, Deka, R, Norgard, M.V. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Inhibition of bacterial FMN transferase: A potential avenue for countering antimicrobial resistance.
Protein Sci., 31, 2022
|
|
4DA1
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase phosphatase with Mg (II) ions at the active site | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Protein phosphatase 1K, ... | | Authors: | Brautigam, C.A, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Structural and biochemical characterization of human mitochondrial branched-chain alpha-ketoacid dehydrogenase phosphatase.
J.Biol.Chem., 287, 2012
|
|
4DI3
 
 | |
4DI4
 
 | | Crystal structure of a 3:1 complex of Treponema pallidum TatP(T) (Tp0957) bound to TatT (Tp0956) | | Descriptor: | TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, TatP(T) (Tp0957), ... | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-01-30 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.714 Å) | | Cite: | Structural and Thermodynamic Characterization of the Interaction between Two Periplasmic Treponema pallidum Lipoproteins that are Components of a TPR-Protein-Associated TRAP Transporter (TPAT).
J.Mol.Biol., 420, 2012
|
|
1PX2
 
 | | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP (Form 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Synapsin I | | Authors: | Brautigam, C.A, Chelliah, Y, Deisenhofer, J. | | Deposit date: | 2003-07-02 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Tetramerization and ATP binding by a protein comprising the A, B, and C domains of rat synapsin I.
J.Biol.Chem., 279, 2004
|
|
1PK8
 
 | | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Brautigam, C.A, Chelliah, Y, Deisenhofer, J. | | Deposit date: | 2003-06-05 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetramerization and ATP binding by a protein comprising the A, B, and C domains of rat synapsin I.
J.Biol.Chem., 279, 2004
|
|
1KFS
 
 | | DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*AP*CP*G)-3'), MAGNESIUM ION, PROTEIN (DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7)), ... | | Authors: | Brautigam, C.A, Steitz, T.A. | | Deposit date: | 1997-08-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural principles for the inhibition of the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates.
J.Mol.Biol., 277, 1998
|
|
1KRP
 
 | | DNA polymerase I Klenow fragment (E.C.2.7.7.7) mutant/DNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*PST)-3'), PROTEIN (DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7)), ZINC ION | | Authors: | Brautigam, C.A, Steitz, T.A. | | Deposit date: | 1997-08-19 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural principles for the inhibition of the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates.
J.Mol.Biol., 277, 1998
|
|
1KSP
 
 | | DNA polymerase I Klenow fragment (E.C.2.7.7.7) mutant/DNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*PST)-3'), PROTEIN (DNA POLYMERASE I-KLENOW FRAGMENT (E.C.2.7.7.7)), ZINC ION | | Authors: | Brautigam, C.A, Steitz, T.A. | | Deposit date: | 1997-08-19 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural principles for the inhibition of the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates.
J.Mol.Biol., 277, 1998
|
|
7JP2
 
 | | Crystal structure of TP0037 from Treponema pallidum, a D-lactate dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-lactate dehydrogenase | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Biophysical and Biochemical Characterization of TP0037, a d-Lactate Dehydrogenase, Supports an Acetogenic Energy Conservation Pathway in Treponema pallidum.
Mbio, 11, 2020
|
|
2KFZ
 
 | | KLENOW FRAGMENT WITH BRIDGING-SULFUR SUBSTRATE AND ZINC ONLY | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*(US1)P*G)-3', KLENOW FRAGMENT OF DNA POLYMERASE I, MAGNESIUM ION, ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-02 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2KFN
 
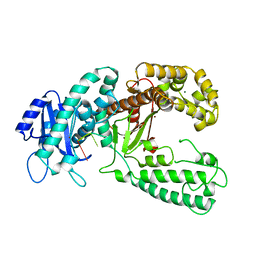 | | KLENOW FRAGMENT WITH BRIDGING-SULFUR SUBSTRATE AND MANGANESE | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*(US1)P*G)-3', KLENOW FRAGMENT OF DNA POLYMERASE I, MAGNESIUM ION, ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-01 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2KZZ
 
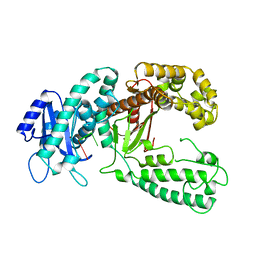 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC ONLY | | Descriptor: | DNA (5'-D(*GP*CP*TP*T*AP*CP*G)-3'), PROTEIN (DNA POLYMERASE I), ZINC ION | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-07 | | Release date: | 1999-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2KZM
 
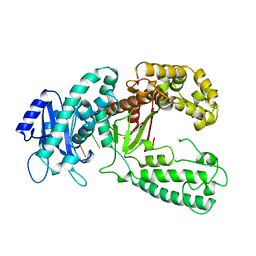 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC AND MANGANESE | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*A*CP*GP*C)-3'), MANGANESE (II) ION, PROTEIN (DNA POLYMERASE I), ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-03 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
3PJN
 
 | | The crystal structure of Tp34 bound to Zn(II) ion at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, SULFATE ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Machius, M, Norgard, M.V. | | Deposit date: | 2010-11-10 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of the Tp34 lipoprotein from Treponema pallidum suggests a role in transition metal homeostasis
To be Published
|
|
3PJL
 
 | | The crystal structure of Tp34 bound to Co (II) ion at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, COBALT (II) ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Machius, M, Norgard, M.V. | | Deposit date: | 2010-11-10 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of the Tp34 lipoprotein from Treponema pallidum suggests a role in transition metal homeostasis
To be Published
|
|
3Q6S
 
 | |
1U3C
 
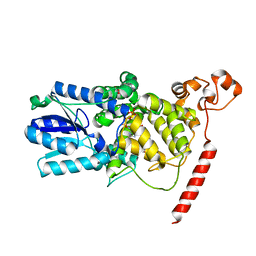 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U3D
 
 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana with AMPPNP bound | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6BGC
 
 | | The crystal structure of the W145A variant of TpMglB-2 (Tp0684) with bound glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucose/galactose-binding lipoprotein, ... | | Authors: | Brautigam, C.A, Norgard, M.V, Deka, R.K. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.082 Å) | | Cite: | Crystal structures of MglB-2 (TP0684), a topologically variant d-glucose-binding protein from Treponema pallidum, reveal a ligand-induced conformational change.
Protein Sci., 27, 2018
|
|
