4DBH
 
 | | Crystal structure of Cg1458 with inhibitor | | Descriptor: | 2-HYDROXYHEPTA-2,4-DIENE-1,7-DIOATE ISOMERASE, MAGNESIUM ION, OXALATE ION | | Authors: | Ran, T.T, Wang, W.W, Xu, D.Q, Gao, Y.Y. | | Deposit date: | 2012-01-15 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of Cg1458 reveal a catalytic lid domain and a common catalytic mechanism for FAH family.
Biochem.J., 449, 2013
|
|
4DBF
 
 | | Crystal structures of Cg1458 | | Descriptor: | 2-HYDROXYHEPTA-2,4-DIENE-1,7-DIOATE ISOMERASE, MAGNESIUM ION | | Authors: | Ran, T.T, Xu, D.Q, Wang, W.W, Gao, Y.Y, Wang, M.T. | | Deposit date: | 2012-01-15 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Cg1458 reveal a catalytic lid domain and a common catalytic mechanism for FAH family.
Biochem.J., 449, 2013
|
|
3J9S
 
 | |
3JWE
 
 | | Crystal structure of human mono-glyceride lipase in complex with SAR629 | | Descriptor: | 1-[bis(4-fluorophenyl)methyl]-4-(1H-1,2,4-triazol-1-ylcarbonyl)piperazine, MGLL protein | | Authors: | Bertrand, T, Auge, F, Houtmann, J, Rak, A, Vallee, F, Mikol, V, Berne, P.F, Michot, N, Cheuret, D, Hoornaert, C, Mathieu, M. | | Deposit date: | 2009-09-18 | | Release date: | 2010-01-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for human monoglyceride lipase inhibition.
J.Mol.Biol., 396, 2010
|
|
6OB3
 
 | | Crystal structure of G13D-KRAS (GMPPNP-bound) in complex with GAP-related domain (GRD) of neurofibromin (NF1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GTPase KRas, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5LX3
 
 | | CRYSTAL STRUCTURE OF VISFATIN IN COMPLEX WITH SAR154782. | | Descriptor: | 6-[4-[(6-azanylpyridin-3-yl)methylcarbamoylamino]-3-fluoranyl-phenyl]-2-(ethylamino)-~{N}-(2-piperidin-1-ylethyl)pyridine-3-carboxamide, Nicotinamide phosphoribosyltransferase | | Authors: | Bertrand, T, Marquette, J.P. | | Deposit date: | 2016-09-20 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF VISFATIN IN COMPLEX WITH SAR154782.
To Be Published
|
|
2PLB
 
 | | D(GTATACC) under hydrostatic pressure of 1.39 GPa | | Descriptor: | 5'-D(*DGP*DGP*DTP*DAP*DTP*DAP*DCP*DC)-3', SPERMINE | | Authors: | Prange, T, Girard, E, Kahn, R, Fourme, R. | | Deposit date: | 2007-04-19 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Adaptation of the base-paired double-helix molecular architecture to extreme pressure.
Nucleic Acids Res., 35, 2007
|
|
3AUL
 
 | | Crystal structure of wild-type Lys48-linked diubiquitin in an open conformation | | Descriptor: | Polyubiquitin-C | | Authors: | Hirano, T, Olivier, S, Yagi, M, Takemoto, E, Hiromoto, T, Satoh, T, Mizushima, T, Kato, K. | | Deposit date: | 2011-02-09 | | Release date: | 2011-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Conformational dynamics of wild-type Lys48-linked diubiquitin in solution
J.Biol.Chem., 286, 2011
|
|
1KYA
 
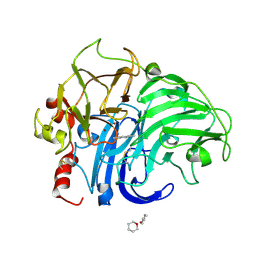 | | ACTIVE LACCASE FROM TRAMETES VERSICOLOR COMPLEXED WITH 2,5-XYLIDINE | | Descriptor: | 2,5-DIMETHYLANILINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Bertrand, T, Jolivalt, C, Briozzo, P, Caminade, E, Joly, N, Madzak, C, Mougin, C. | | Deposit date: | 2002-02-04 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a four-copper laccase complexed with an arylamine: insights into substrate recognition and correlation with kinetics.
Biochemistry, 41, 2002
|
|
1LB3
 
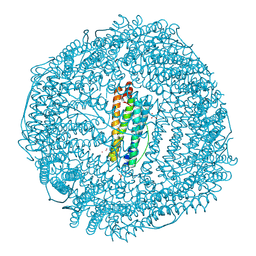 | | Structure of recombinant mouse L chain ferritin at 1.2 A resolution | | Descriptor: | CADMIUM ION, FERRITIN LIGHT CHAIN 1, GLYCEROL, ... | | Authors: | Granier, T, Langlois D'Estaintot, B, Gallois, B, Chevalier, J.-M, Precigoux, G, Santambrogio, P, Arosio, P. | | Deposit date: | 2002-04-02 | | Release date: | 2003-01-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural description of the active sites of mouse L-chain ferritin at 1.2A resolution
J.Biol.Inorg.Chem., 8, 2003
|
|
4KNF
 
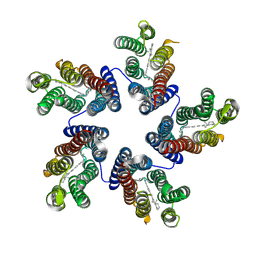 | | Crystal structure of a blue-light absorbing proteorhodopsin double-mutant D97N/Q105L from HOT75 | | Descriptor: | Blue-light absorbing proteorhodopsin, RETINAL | | Authors: | Ran, T, Ozorowski, G, Gao, Y, Wang, W, Luecke, H. | | Deposit date: | 2013-05-09 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KLY
 
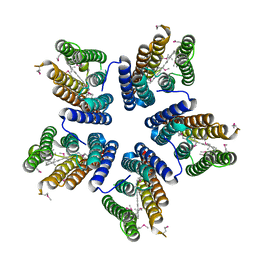 | | Crystal structure of a blue-light absorbing proteorhodopsin mutant D97N from HOT75 | | Descriptor: | Blue-light absorbing proteorhodopsin, RETINAL | | Authors: | Ran, T, Ozorowski, G, Gao, Y, Wang, W, Luecke, H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6AEO
 
 | | TssL periplasmic domain | | Descriptor: | GLYCEROL, Maltose/maltodextrin-binding periplasmic protein,TssL | | Authors: | Ran, T.T, Wang, W.W, Wang, X.B, Xu, D.Q. | | Deposit date: | 2018-08-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the periplasmic domain of TssL, a key membrane component of Type VI secretion system.
Int.J.Biol.Macromol., 120, 2018
|
|
8GQR
 
 | | Crystal structure of VioD with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Ran, T, Wang, W, Xu, M. | | Deposit date: | 2022-08-30 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of the key enzyme VioD in the violacein synthesis pathway.
Proteins, 91, 2023
|
|
5YLT
 
 | | Crystal structure of SET7/9 in complex with a cyproheptadine derivative | | Descriptor: | 2-(1-methylpiperidin-4-ylidene)tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3(8),4,6,9,12,14-heptaen-6-ol, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Hirano, T, Fujiwara, T, Niwa, H, Hirano, M, Ohira, K, Okazaki, Y, Sato, S, Umehara, T, Maemoto, Y, Ito, A, Yoshida, M, Kagechika, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of Novel Inhibitors for Histone Methyltransferase SET7/9 based on Cyproheptadine.
ChemMedChem, 13, 2018
|
|
6WS2
 
 | |
5MJD
 
 | | metNgb under oxygen at 80 bar | | Descriptor: | Neuroglobin, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
5MJC
 
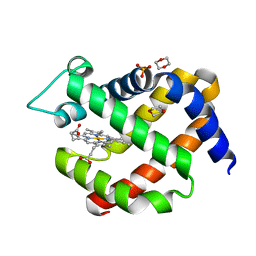 | | metNeuroglobin under oxygen at 50 bar | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Neuroglobin, OXYGEN MOLECULE, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P, Vallone, B. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
6XCO
 
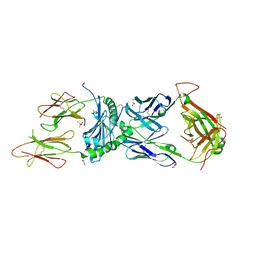 | | Immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, GLYCEROL, ... | | Authors: | Tran, T.M, Faridi, P, Lim, J.J, Ting, T.Y, Onwukwe, G, Bhattacharjee, P, Jones, M.C, Tresoldi, E, Cameron, J.F, La-Gruta, L.N, Purcell, W.A, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2020-06-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | T cell receptor recognition of hybrid insulin peptides bound to HLA-DQ8.
Nat Commun, 12, 2021
|
|
6XC9
 
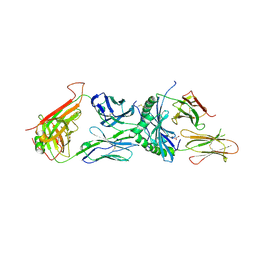 | | Immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Tran, T.M, Faridi, P, Lim, J.J, Ting, T.Y, Onwukwe, G, Bhattacharjee, P, Tresoldi, M.C.E, Cameron, J.F, La-Gruta, L.N, Purcell, W.A, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2020-06-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | T cell receptor recognition of hybrid insulin peptides bound to HLA-DQ8.
Nat Commun, 12, 2021
|
|
6XCP
 
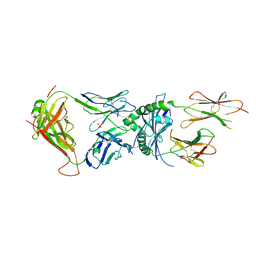 | | Immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hybrid insulin peptide, ... | | Authors: | Tran, T.M, Faridi, P, Lim, J.J, Ting, T.Y, Onwukwe, G, Bhattacharjee, P, Jones, M.C, Tresoldi, E, Cameron, J.F, La-Gruta, L.N, Purcell, W.A, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2020-06-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | T cell receptor recognition of hybrid insulin peptides bound to HLA-DQ8.
Nat Commun, 12, 2021
|
|
4RCN
 
 | |
6WS4
 
 | | Crystal structure of KRAS-G12D/K104Q mutant, GDP-bound | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tran, T.H, Simanshu, D.K. | | Deposit date: | 2020-04-30 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Allosteric regulation of switch-II domain controls KRAS oncogenicity.
Cancer Res., 2023
|
|
7PUF
 
 | | urate oxidase azaxanthine complex under 600 bar (60 MPa) of argon | | Descriptor: | 8-AZAXANTHINE, ARGON, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
7A47
 
 | | KRASG12C GDP form in complex with Cpd4 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ~{N}-[3-bromanyl-2-(2-methylimidazol-1-yl)pyridin-4-yl]-3-[[3-bromanyl-2-(2-methylimidazol-1-yl)pyridin-4-yl]-propanoyl-amino]propanamide | | Authors: | Bertrand, T, Steier, V. | | Deposit date: | 2020-08-19 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | KRAS G12C fragment screening renders new binding pockets.
Small Gtpases, 13, 2022
|
|
