8VKK
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | Descriptor: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBO
 
 | | Structure of HEPATITIS C VIRAL NS3 protease domain complexed with NS4A peptide and ketoamide SCH476776 | | Descriptor: | BETA-MERCAPTOETHANOL, HCV NS3 protease, HCV NS4A peptide, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
6X1Q
 
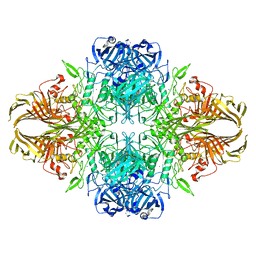 | | 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope | | Descriptor: | Beta-galactosidase, MAGNESIUM ION, SODIUM ION | | Authors: | Merk, A, Fukumura, T, Zhu, X, Darling, J, Grisshammer, R, Ognjenovic, J, Subramaniam, S. | | Deposit date: | 2020-05-19 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | 1.8 angstrom resolution structure of beta-galactosidase with a 200 kV CRYO ARM electron microscope.
Iucrj, 7, 2020
|
|
6CMO
 
 | | Rhodopsin-Gi complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | Authors: | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
6PWF
 
 | |
6OSI
 
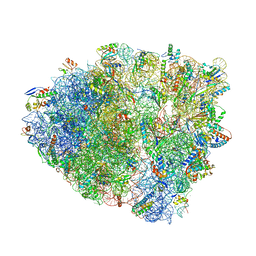 | | Unmodified tRNA(Pro) bound to Thermus thermophilus 70S (near cognate) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Subaramanian, S, Hong, S, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2019-05-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4.14 Å) | | Cite: | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
6PWE
 
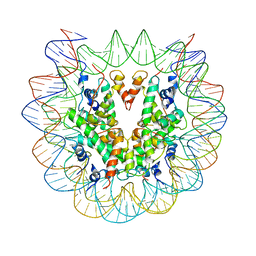 | | Cryo-EM structure of nucleosome core particle | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B, ... | | Authors: | Chittori, S, Subramaniam, S. | | Deposit date: | 2019-07-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structure of the primed state of the ATPase domain of chromatin remodeling factor ISWI bound to the nucleosome.
Nucleic Acids Res., 47, 2019
|
|
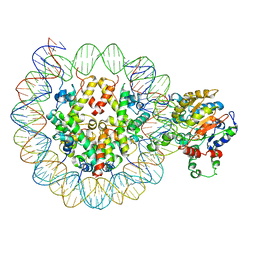
6UAN
 
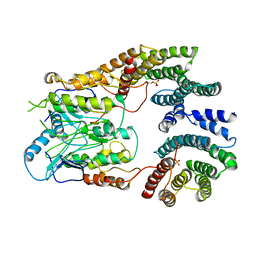 | | B-Raf:14-3-3 complex | | Descriptor: | 14-3-3 zeta, Serine/threonine-protein kinase B-raf | | Authors: | Kondo, Y, Ognjenovic, J, Banerjee, S, Karandur, D, Merk, A, Kulhanek, K, Wong, K, Roose, J.P, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases.
Science, 366, 2019
|
|
7SZ7
 
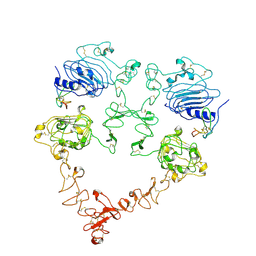 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to TGF-alpha. "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor receptor, Transforming growth factor alpha | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SZ1
 
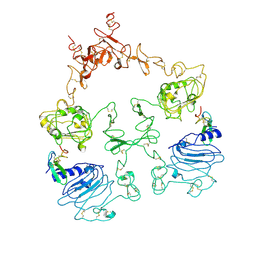 | | Cryo-EM structure of the extracellular module of the full-length EGFR L834R bound to EGF. "tips-separated" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SZ0
 
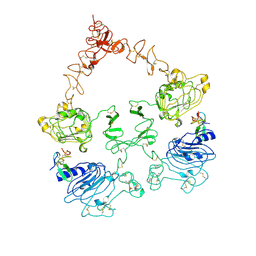 | | Cryo-EM structure of the extracellular module of the full-length EGFR L834R bound to EGF. "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SYD
 
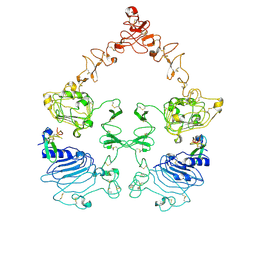 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SZ5
 
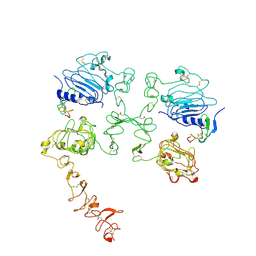 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to TGF-alpha "tips-separated" conformation | | Descriptor: | Epidermal growth factor receptor, Transforming growth factor alpha | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SYE
 
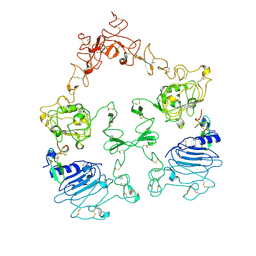 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF. "tips-separated" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7T9L
 
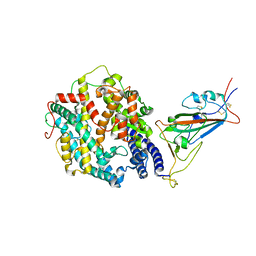 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
7T9J
 
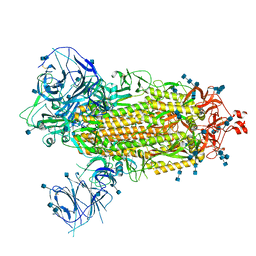 | | Cryo-EM structure of the SARS-CoV-2 Omicron spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
7T9K
 
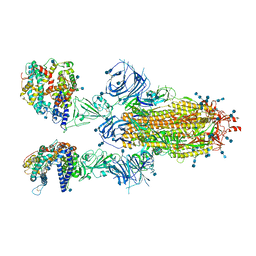 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
6CGV
 
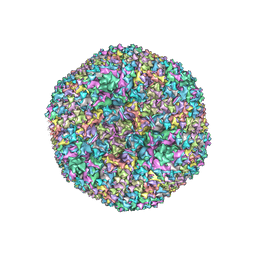 | | Revised crystal structure of human adenovirus | | Descriptor: | Hexon protein, Hexon-interlacing protein, Penton protein, ... | | Authors: | Natchiar, S.K, Venkataraman, S, Nemerow, G.R, Reddy, V.S. | | Deposit date: | 2018-02-21 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Revised Crystal Structure of Human Adenovirus Reveals the Limits on Protein IX Quasi-Equivalence and on Analyzing Large Macromolecular Complexes.
J. Mol. Biol., 430, 2018
|
|
7TEW
 
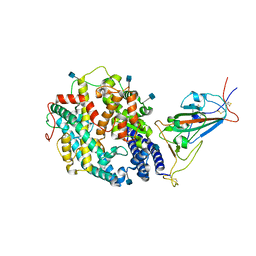 | | Cryo-EM structure of SARS-CoV-2 Delta (B.1.617.2) spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
7TEY
 
 | | Cryo-EM structure of SARS-CoV-2 Delta (B.1.617.2) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
7TEZ
 
 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
7TF2
 
 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) Q484I spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
