5OO1
 
 | |
5OQA
 
 | |
6XXG
 
 | | Structure of truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Deinococcus radiodurans | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate synthase, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Gierse, R.M, Reddem, E, Grooves, M.R. | | Deposit date: | 2020-01-27 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Identification of a 1-deoxy-D-xylulose-5-phosphate synthase (DXS) mutant with improved crystallographic properties.
Biochem.Biophys.Res.Commun., 539, 2021
|
|
3I3I
 
 | |
3I03
 
 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) - monomeric form at a high resolution | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
6XTF
 
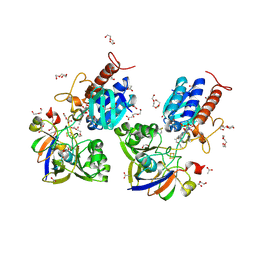 | | Crystal structure a Thioredoxin Reductase from Gloeobacter violaceus bound to its electron donor | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Buey, R.M, Gonzalez-Holgado, G, Fernandez-Justel, D, Balsera, M. | | Deposit date: | 2020-01-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Unexpected diversity of ferredoxin-dependent thioredoxin reductases in cyanobacteria.
Plant Physiol., 186, 2021
|
|
3IQ3
 
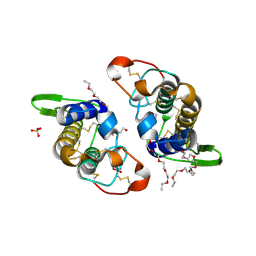 | | Crystal Structure of Bothropstoxin-I complexed with polietilene glicol 4000 - crystallized at 283 K | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Phospholipase A2 homolog bothropstoxin-1, SULFATE ION | | Authors: | Salvador, G.H.M, Marchi-Salvador, D.P, Silva, M.C.O, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-08-19 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
5MKK
 
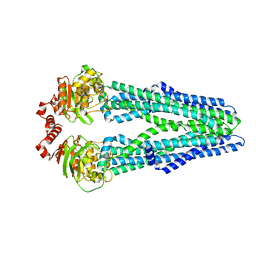 | | Crystal structure of the heterodimeric ABC transporter TmrAB, a homolog of the antigen translocation complex TAP | | Descriptor: | Multidrug resistance ABC transporter ATP-binding and permease protein, SULFATE ION | | Authors: | Noell, A, Thomas, C, Tomasiak, T.M, Olieric, V, Wang, M, Diederichs, K, Stroud, R.M, Pos, K.M, Tampe, R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and mechanistic basis of a functional homolog of the antigen transporter TAP.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6Y5B
 
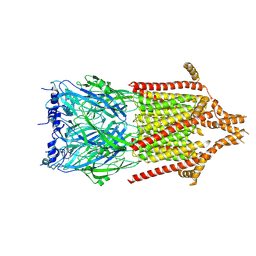 | | 5-HT3A receptor in Salipro (apo, asymmetric) | | Descriptor: | 5-hydroxytryptamine receptor 3A | | Authors: | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | Deposit date: | 2020-02-25 | | Release date: | 2020-12-23 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
3ICD
 
 | | STRUCTURE OF A BACTERIAL ENZYME REGULATED BY PHOSPHORYLATION, ISOCITRATE DEHYDROGENASE | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Thorsness, P.E, Ramalingam, V, Helmers, N.H, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1989-12-28 | | Release date: | 1991-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 86, 1989
|
|
3GEN
 
 | | The 1.6 A crystal structure of human bruton's tyrosine kinase bound to a pyrrolopyrimidine-containing compound | | Descriptor: | 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane, Tyrosine-protein kinase BTK | | Authors: | Marcotte, D.J, Liu, Y.T, Arduini, R.M, Hession, C.A, Miatkowski, K, Wildes, C.P, Cullen, P.F, Hopkins, B, Mertsching, E, Jenkins, T.J, Romanowski, M.J, Baker, D.P, Silvian, L.F. | | Deposit date: | 2009-02-25 | | Release date: | 2010-01-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human Bruton's tyrosine kinase in active and inactive conformations suggest a mechanism of activation for TEC family kinases.
Protein Sci., 19, 2010
|
|
5MZP
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with caffeine at 2.1A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CAFFEINE, ... | | Authors: | Cheng, K.Y.R, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
5NDD
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ8838 at 2.8 angstrom resolution | | Descriptor: | (~{S})-(4-fluoranyl-2-propyl-phenyl)-(1~{H}-imidazol-2-yl)methanol, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, PHOSPHATE ION, ... | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
3GH3
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, Ecto-NAD+ glycohydrolase (CD38 molecule), ... | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Kellenberger, E, Oppenheimer, N, Schuber, F. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
3GF1
 
 | |
3K54
 
 | | Structures of human Bruton's tyrosine kinase in active and inactive conformations suggests a mechanism of activation for TEC family kinases. | | Descriptor: | N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, Tyrosine-protein kinase BTK | | Authors: | Marcotte, D.J, Liu, Y.-T, Arduini, R.M, Hession, C.A, Miatkowski, K, Wildes, C.P, Cullen, P.F, Hopkins, B.T, Mertsching, E, Jenkins, T.J, Romanowski, M.J, Baker, D.P, Silvian, L.F. | | Deposit date: | 2009-10-06 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of human Bruton's tyrosine kinase in active and inactive conformations suggest a mechanism of activation for TEC family kinases.
Protein Sci., 19, 2010
|
|
5OOZ
 
 | |
5OQE
 
 | |
3JAN
 
 | |
5OQ9
 
 | |
5OSJ
 
 | | Cdk2(WT) with covalent adduct at C177 | | Descriptor: | Cyclin-dependent kinase 2, ~{tert}-butyl 4-propanoyl-2,3-dihydroquinoxaline-1-carboxylate | | Authors: | Craven, G, Morgan, R.M.L, Mann, D.J. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | High-Throughput Kinetic Analysis for Target-Directed Covalent Ligand Discovery.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3HRC
 
 | |
5OO3
 
 | |
5OSM
 
 | | Cdk2(F80C, C177A) with covalent adduct at C80 | | Descriptor: | Cyclin-dependent kinase 2, methyl 1-propanoyl-3,4-dihydro-2~{H}-quinoline-6-carboxylate | | Authors: | Craven, G, Morgan, R.M.L, Mann, D.J. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | High-Throughput Kinetic Analysis for Target-Directed Covalent Ligand Discovery.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3K8N
 
 | |
