6J10
 
 | | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly | | Descriptor: | 6-cyclohexyl-4-methyl-1-oxidanyl-pyridin-2-one, Capsid protein | | Authors: | Park, S, Jin, M.S, Cho, Y, Kang, J, Kim, S, Park, M, Park, H, Kim, J, Park, S, Hwang, J, Kim, Y, Kim, Y.J. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly.
Nat Commun, 10, 2019
|
|
1CMJ
 
 | |
1CL6
 
 | |
1CMN
 
 | |
5CSS
 
 | | Crystal structure of triosephosphate isomerase from Thermoplasma acidophilum with glycerol 3-phosphate | | Descriptor: | CHLORIDE ION, SN-GLYCEROL-3-PHOSPHATE, Triosephosphate isomerase | | Authors: | Park, S.H, Kim, H.S, Song, M.K, Kim, K.R, Park, J.S, Han, B.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Stability of the Dimeric Triosephosphate Isomerase from the Thermophilic Archaeon Thermoplasma acidophilum.
Plos One, 10, 2015
|
|
5CSR
 
 | | Crystal structure of triosephosphate isomerase from Thermoplasma acidophilium | | Descriptor: | CHLORIDE ION, GLYCEROL, Triosephosphate isomerase | | Authors: | Park, S.H, Kim, H.S, Song, M.K, Park, H.S, Han, B.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure and Stability of the Dimeric Triosephosphate Isomerase from the Thermophilic Archaeon Thermoplasma acidophilum.
Plos One, 10, 2015
|
|
4N7Z
 
 | | Crystal structure of human Plk4 cryptic polo box (CPB) in complex with a Cep192 N-terminal fragment | | Descriptor: | Centrosomal protein of 192 kDa, Serine/threonine-protein kinase PLK4 | | Authors: | Park, S.-Y, Park, J.-E, Tian, L, Kim, T.-S, Yang, W, Lee, K.S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5GV5
 
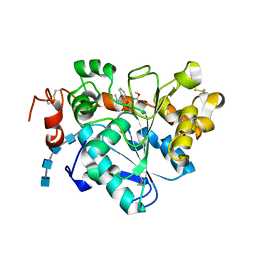 | | Crystal structure of Candida antarctica Lipase B with active Ser105 modified with a phosphonate inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B, ... | | Authors: | Park, S.Y, Lee, H. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural and Experimental Evidence for the Enantiomeric Recognition toward a Bulky sec-Alcohol by Candida antarctica Lipase B
Acs Catalysis, 6, 2016
|
|
6W5N
 
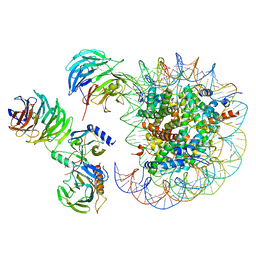 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class05) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5M
 
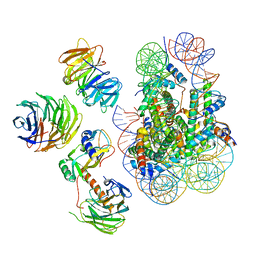 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class02) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5I
 
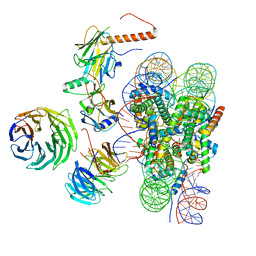 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class01) | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
8EY0
 
 | | Structure of an orthogonal PYR1*:HAB1* chemical-induced dimerization module in complex with mandipropamid | | Descriptor: | (2S)-2-(4-chlorophenyl)-N-{2-[3-methoxy-4-(prop-2-yn-1-yloxy)phenyl]ethyl}-2-(prop-2-yn-1-yloxy)ethanamide, Abscisic acid receptor PYR1, GLYCEROL, ... | | Authors: | Park, S.-Y, Volkman, B.F, Cutler, S.R, Peterson, F.C. | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An orthogonalized PYR1-based CID module with reprogrammable ligand-binding specificity.
Nat.Chem.Biol., 20, 2024
|
|
8KCA
 
 | |
6PWW
 
 | | Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6PWV
 
 | | Cryo-EM structure of MLL1 core complex bound to the nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
4YUS
 
 | | Crystal structure of photoactivated adenylyl cyclase of a cyanobacteriaOscillatoria acuminata in hexagonal form | | Descriptor: | FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase | | Authors: | Park, S.-Y, Ohki, M, Sugiyama, K, Kawai, F, Iseki, M. | | Deposit date: | 2015-03-19 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into photoactivation of an adenylate cyclase from a photosynthetic cyanobacterium
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YUT
 
 | | Crystal structure of photoactivated adenylyl cyclase of a cyanobacteriaOscillatoria acuminata in orthorhombic form | | Descriptor: | FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase | | Authors: | Park, S.-Y, Ohki, M, Sugiyama, K, Kawai, F, Iseki, M. | | Deposit date: | 2015-03-19 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into photoactivation of an adenylate cyclase from a photosynthetic cyanobacterium
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7XL9
 
 | | The structure of HucR with urate | | Descriptor: | CHLORIDE ION, Transcriptional regulator, MarR family, ... | | Authors: | Park, S.Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The structure of Deinococcus radiodurans transcriptional regulator HucR retold with the urate bound.
Biochem.Biophys.Res.Commun., 615, 2022
|
|
6JLD
 
 | | Crystal structure of a human ependymin related protein | | Descriptor: | Mammalian ependymin-related protein 1 | | Authors: | Park, S.Y. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of three ependymin-related proteins suggest their function as a hydrophobic molecule binder.
Iucrj, 6, 2019
|
|
6JL9
 
 | | Crystal structure of a frog ependymin related protein | | Descriptor: | CALCIUM ION, Ependymin-related 1 | | Authors: | Park, S.Y. | | Deposit date: | 2019-03-04 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of three ependymin-related proteins suggest their function as a hydrophobic molecule binder.
Iucrj, 6, 2019
|
|
6L18
 
 | |
6PWX
 
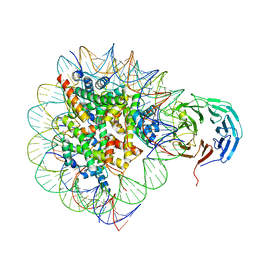 | | Cryo-EM structure of RbBP5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
4YWV
 
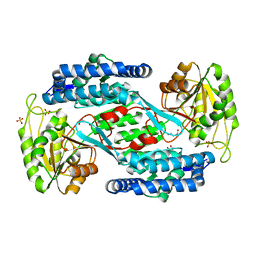 | | Structural insight into the substrate inhibition mechanism of NADP+-dependent succinic semialdehyde dehydrogenase from Streptococcus pyogenes | | Descriptor: | 4-oxobutanoic acid, SULFATE ION, Succinic semialdehyde dehydrogenase | | Authors: | Park, S.A, Jang, E.H, Chi, Y.M, Lee, K.S. | | Deposit date: | 2015-03-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the substrate inhibition mechanism of NADP(+)-dependent succinic semialdehyde dehydrogenase from Streptococcus pyogenes.
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4N7V
 
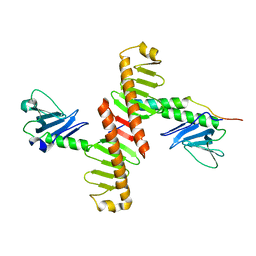 | | Crystal structure of human Plk4 cryptic polo box (CPB) in complex with a Cep152 N-terminal fragment | | Descriptor: | Centrosomal protein of 152 kDa, Serine/threonine-protein kinase PLK4 | | Authors: | Park, S.-Y, Park, J.-E, Tian, L, Kim, T.-S, Yang, W, Lee, K.S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat.Struct.Mol.Biol., 21, 2014
|
|
8SQG
 
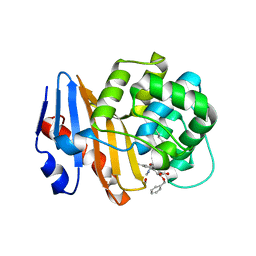 | | OXA-48 bound to inhibitor CDD-2801 | | Descriptor: | (1M)-3'-(benzyloxy)-5-[2-(methylamino)-2-oxoethoxy][1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
