1OQJ
 
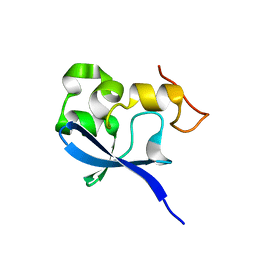 | | Crystal structure of the SAND domain from glucocorticoid modulatory element binding protein-1 (GMEB1) | | Descriptor: | Glucocorticoid Modulatory Element Binding protein-1, ZINC ION | | Authors: | Surdo, P.L, Bottomley, M.J, Sattler, M, Scheffzek, K. | | Deposit date: | 2003-03-10 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and nuclear magnetic resonance analyses of the SAND domain from glucocorticoid modulatory element binding protein-1 reveals deoxyribonucleic acid and zinc binding regions
MOL.ENDOCRINOL., 17, 2003
|
|
1NC3
 
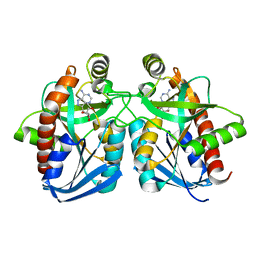 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with formycin A (FMA) | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2002-12-04 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
1S4N
 
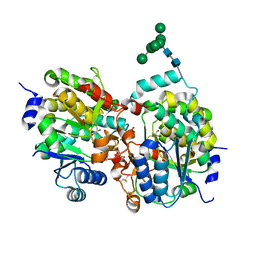 | | Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lobsanov, Y.D, Romero, P.A, Sleno, B, Yu, B, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 2004-01-16 | | Release date: | 2004-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of Kre2p/Mnt1p: A YEAST {alpha}1,2-MANNOSYLTRANSFERASE INVOLVED IN MANNOPROTEIN BIOSYNTHESIS
J.Biol.Chem., 279, 2004
|
|
1S4O
 
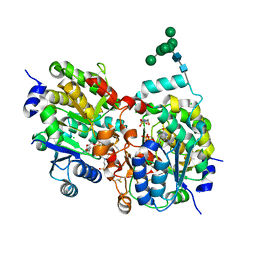 | | Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p: binary complex with GDP/Mn | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Lobsanov, Y.D, Romero, P.A, Sleno, B, Yu, B, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 2004-01-16 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of Kre2p/Mnt1p: A YEAST {alpha}1,2-MANNOSYLTRANSFERASE INVOLVED IN MANNOPROTEIN BIOSYNTHESIS
J.Biol.Chem., 279, 2004
|
|
1OBN
 
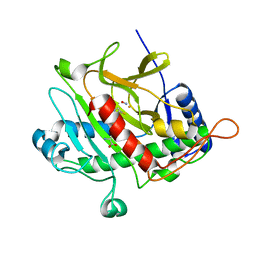 | | ISOPENICILLIN N SYNTHASE aminoadipoyl-cysteinyl-aminobutyrate-FE-NO COMPLEX | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-D-VINYLGLYCINE, FE (II) ION, HYDROXYAMINE, ... | | Authors: | Long, A.J, Clifton, I.J, Roach, P.L, Baldwin, J.E, Schofield, C.J, Rutledge, P.J. | | Deposit date: | 2003-01-31 | | Release date: | 2004-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with the Substrate Analogue Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-D-Alpha-Aminobutyrate.
Biochem.J., 372, 2003
|
|
1ODN
 
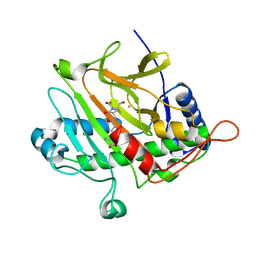 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (OXYGEN-EXPOSED PRODUCT FROM ANAEROBIC AC-VINYLGLYCINE FE COMPLEX) | | Descriptor: | 6-(5-AMINO-5-CARBOXY-PENTANOYLAMINO)-3-HYDROXYMETHYL-7-OXO-4-THIA-1-AZA-BICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Elkins, J.M, Rutledge, P.J, Burzlaff, N.I, Clifton, I.J, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2003-02-19 | | Release date: | 2003-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Studies on the Reaction of Isopenicillin N Synthase with an Unsaturated Substrate Analogue
Org.Biomol.Chem., 1, 2003
|
|
1RGQ
 
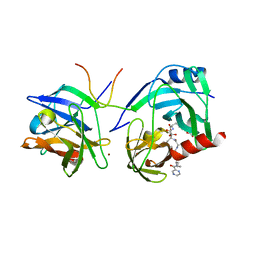 | | M9A HCV Protease complex with pentapeptide keto-amide inhibitor | | Descriptor: | N-(PYRAZIN-2-YLCARBONYL)LEUCYLISOLEUCYL-N~1~-{1-[2-({1-CARBOXY-2-[4-(PHOSPHONOOXY)PHENYL]ETHYL}AMINO)-1,1-DIHYDROXY-2-OXOETHYL]BUT-3-ENYL}-3-CYCLOHEXYLALANINAMIDE, NS3 Protease, NS4A peptide, ... | | Authors: | Liu, Y, Stoll, V.S, Richardson, P.L, Saldivar, A, Klaus, J.L, Molla, A, Kohlbrenner, W, Kati, W.M. | | Deposit date: | 2003-11-12 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Hepatitis C NS3 protease inhibition by peptidyl-alpha-ketoamide inhibitors: kinetic mechanism and structure.
Arch.Biochem.Biophys., 421, 2004
|
|
1U16
 
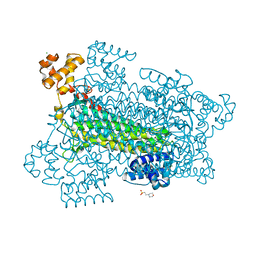 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) in complex with sulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Delta crystallin I, ... | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
1U15
 
 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) | | Descriptor: | Delta crystallin I | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
1NH5
 
 | | AUTOMATIC ASSIGNMENT OF NMR DATA AND DETERMINATION OF THE PROTEIN STRUCTURE OF A NEW WORLD SCORPION NEUROTOXIN USING NOAH/DIAMOD | | Descriptor: | Neurotoxin 5 | | Authors: | Xu, Y, Jablonsky, M.J, Jackson, P.L, Krishna, N.R, Braun, W. | | Deposit date: | 2002-12-18 | | Release date: | 2003-01-07 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Automatic assignment of NOESY Cross peaks and determination of the protein structure of a new world scorpion neurotoxin Using NOAH/DIAMOD
J.Magn.Reson., 148, 2001
|
|
1P4O
 
 | | Structure of Apo unactivated IGF-1R KInase domain at 1.5A resolution. | | Descriptor: | Insulin-like growth factor I receptor protein | | Authors: | Munshi, S, Kornienko, M, Hall, D.L, Darke, P.L, Waxman, L, Kuo, L.C. | | Deposit date: | 2003-04-23 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of apo, unactivated insulin-like growth factor-1 receptor kinase at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1RM6
 
 | | Structure of 4-hydroxybenzoyl-CoA reductase from Thauera aromatica | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxybenzoyl-CoA reductase alpha subunit, ... | | Authors: | Unciuleac, M, Warkentin, E, Page, C.C, Dutton, P.L, Boll, M, Ermler, U. | | Deposit date: | 2003-11-27 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a Xanthine Oxidase-Related 4-Hydroxybenzoyl-CoA Reductase with an Additional [4Fe-4S] Cluster and an Inverted Electron Flow
Structure, 12, 2004
|
|
1OC1
 
 | | ISOPENICILLIN N SYNTHASE aminoadipoyl-cysteinyl-aminobutyrate-FE COMPLEX | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-D-VINYLGLYCINE, FE (II) ION, ISOPENICILLIN N SYNTHETASE, ... | | Authors: | Long, A.J, Clifton, I.J, Roach, P.L, Baldwin, J.E, Schofield, C.J, Rutledge, P.J. | | Deposit date: | 2003-02-03 | | Release date: | 2004-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with the Substrate Analogue Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-D-Alpha-Aminobutyrate
Biochem.J., 372, 2003
|
|
1U6B
 
 | | CRYSTAL STRUCTURE OF A SELF-SPLICING GROUP I INTRON WITH BOTH EXONS | | Descriptor: | 197-MER, 5'-R(*AP*AP*GP*CP*CP*AP*CP*AP*CP*AP*AP*AP*CP*CP*AP*GP*AP*CP*GP *GP*CP*C)-3', 5'-R(*CP*AP*(5MU))-3', ... | | Authors: | Adams, P.L, Stahley, M.R, Kosek, A.B, Wang, J, Strobel, S.A. | | Deposit date: | 2004-07-29 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Self-Splicing Group I Intron with Both Exons.
Nature, 430, 2004
|
|
1NP8
 
 | | 18-k C-terminally trunucated small subunit of calpain | | Descriptor: | CADMIUM ION, Calcium-dependent protease, small subunit | | Authors: | Leinala, E.K, Arthur, J.S, Grochulski, P, Davies, P.L, Elce, J.S, Jia, Z. | | Deposit date: | 2003-01-17 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A second binding site revealed by C-terminal truncation of calpain small subunit, a penta-EF-hand protein
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
1MSI
 
 | |
1ODM
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ANAEROBIC AC-VINYLGLYCINE FE COMPLEX) | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-D-VINYLGLYCINE, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Elkins, J.M, Rutledge, P.J, Burzlaff, N.I, Clifton, I.J, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2003-02-19 | | Release date: | 2003-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallographic Studies on the Reaction of Isopenicillin N Synthase with an Unsaturated Substrate Analogue
Org.Biomol.Chem., 1, 2003
|
|
1TBW
 
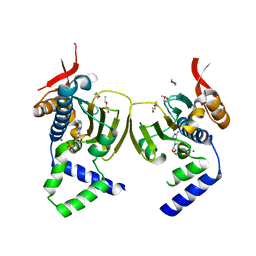 | | Ligand Induced Conformational Shift in the N-terminal Domain of GRP94, Open Conformation | | Descriptor: | ADENOSINE MONOPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
1N4I
 
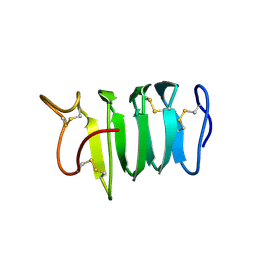 | | Solution structure of spruce budworm antifreeze protein at 5 degrees celsius | | Descriptor: | thermal hysteresis protein | | Authors: | Graether, S.P, Gagne, S.M, Spyracopoulos, L, Jia, Z, Davies, P.L, Sykes, B.D. | | Deposit date: | 2002-10-31 | | Release date: | 2003-04-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Spruce Budworm Antifreeze Protein: Changes in Structure and Dynamics at Low Temperature
J.Mol.Biol., 327, 2003
|
|
1TL9
 
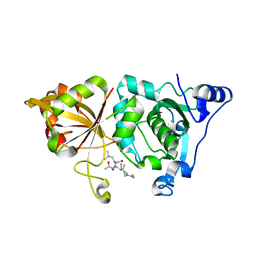 | | High resolution crystal structure of calpain I protease core in complex with leupeptin | | Descriptor: | CALCIUM ION, Calpain 1, large [catalytic] subunit, ... | | Authors: | Moldoveanu, T, Campbell, R.L, Cuerrier, D, Davies, P.L. | | Deposit date: | 2004-06-09 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Calpain-E64 and -Leupeptin Inhibitor Complexes Reveal Mobile Loops Gating the Active Site
J.Mol.Biol., 343, 2004
|
|
1N5Y
 
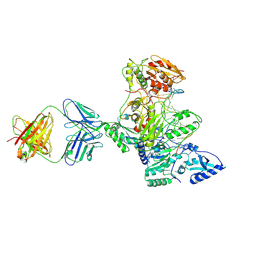 | | HIV-1 Reverse Transcriptase Crosslinked to Post-Translocation AZTMP-Terminated DNA (Complex P) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3', 5'-D(*AP*TP*GP*C*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, P, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of HIV-1 Reverse Transcriptase with Pre-Translocation and Post-Translocation AZTMP-Terminated DNA
Embo J., 21, 2002
|
|
1TC0
 
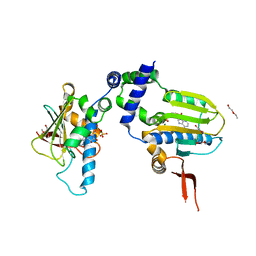 | | Ligand Induced Conformational Shifts in the N-terminal Domain of GRP94, Open Conformation Complexed with the physiological partner ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
1N6Q
 
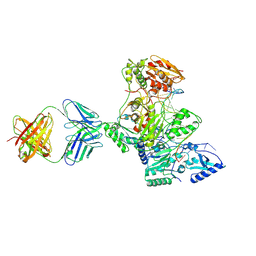 | | HIV-1 Reverse Transcriptase Crosslinked to pre-translocation AZTMP-terminated DNA (complex N) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*AP*(ATM))-3', 5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, I, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-11 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 Reverse Transcriptase with Pre- and Post-translocation AZTMP-terminated DNA
Embo J., 21, 2002
|
|
1MDW
 
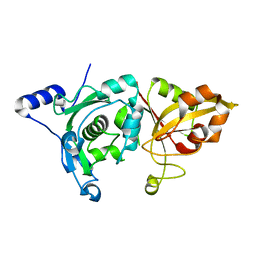 | | Crystal Structure of Calcium-Bound Protease Core of Calpain II Reveals the Basis for Intrinsic Inactivation | | Descriptor: | CALCIUM ION, Calpain II, catalytic subunit | | Authors: | Moldoveanu, T, Hosfield, C.M, Lim, D, Jia, Z, Davies, P.L. | | Deposit date: | 2002-08-07 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Calpain silencing by a reversible intrinsic mechanism.
Nat.Struct.Biol., 10, 2003
|
|
1MJE
 
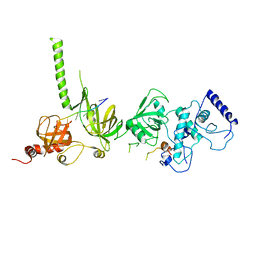 | | STRUCTURE OF A BRCA2-DSS1-SSDNA COMPLEX | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*T)-3', Deleted in split hand/split foot protein 1, breast cancer 2 | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-27 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure.
Science, 297, 2002
|
|
