4XQ8
 
 | |
8GQO
 
 | |
7D0V
 
 | | Crystal structure of Taiwan cobra 5'-nucleotidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Snake venom 5'-nucleotidase, ... | | Authors: | Tsai, M.-H, Lin, I.-J, Lin, C.-C, Wu, W.-G. | | Deposit date: | 2020-09-12 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Crystal structure of Taiwan cobra 5'-nucleotidase
To Be Published
|
|
2PTQ
 
 | |
2PTR
 
 | |
2PTS
 
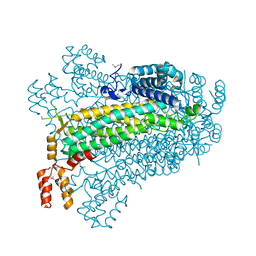 | |
1U16
 
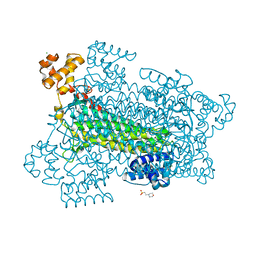 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) in complex with sulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Delta crystallin I, ... | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
1U15
 
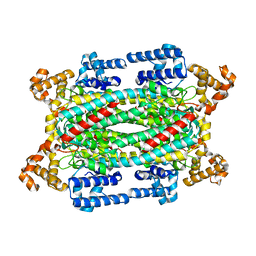 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) | | Descriptor: | Delta crystallin I | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
3O7W
 
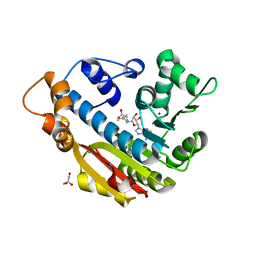 | | The Crystal Structure of Human Leucine Carboxyl Methyltransferase 1 | | Descriptor: | GLYCEROL, Leucine carboxyl methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Tsai, M.L, Cronin, N, Djordjevic, S. | | Deposit date: | 2010-08-01 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of human leucine carboxyl methyltransferase 1 that regulates protein phosphatase PP2A
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1BU9
 
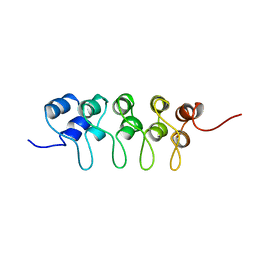 | | SOLUTION STRUCTURE OF P18-INK4C, 21 STRUCTURES | | Descriptor: | PROTEIN (CYCLIN-DEPENDENT KINASE 6 INHIBITOR) | | Authors: | Byeon, I.-J.L, Li, J, Tsai, M.-D. | | Deposit date: | 1998-09-15 | | Release date: | 1999-09-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: determination of the solution structure of p18INK4C and demonstration of the functional significance of loops in p18INK4C and p16INK4A.
Biochemistry, 38, 1999
|
|
1C74
 
 | | Structure of the double mutant (K53,56M) of phospholipase A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sekar, K, Tsai, M.D, Jain, M.K, Ramakumar, S. | | Deposit date: | 2000-01-22 | | Release date: | 2000-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the anionic interface preference and k*cat activation of pancreatic phospholipase A2.
Biochemistry, 39, 2000
|
|
8XMI
 
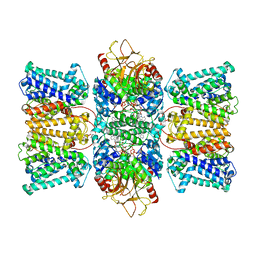 | | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, C1 symmetry | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Chang, Y.K, Chiang, W.T, Hu, N.J, Tsai, M.D. | | Deposit date: | 2023-12-27 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, C1 symmetry
To Be Published
|
|
8XMH
 
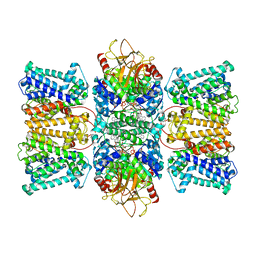 | | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, vertical C2 symmetry axis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Chang, Y.K, Chiang, W.T, Hu, N.J, Tsai, M.D. | | Deposit date: | 2023-12-27 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, vertical C2 symmetry axis
To Be Published
|
|
1BVM
 
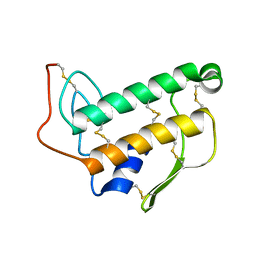 | | SOLUTION NMR STRUCTURE OF BOVINE PANCREATIC PHOSPHOLIPASE A2, 20 STRUCTURES | | Descriptor: | PROTEIN (PHOSPHOLIPASE A2) | | Authors: | Yuan, C.-H, Byeon, I.-J.L, Li, Y, Tsai, M.-D. | | Deposit date: | 1998-09-14 | | Release date: | 1999-09-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of phospholipase A2 from functional perspective. 1. Functionally relevant solution structure and roles of the hydrogen-bonding network.
Biochemistry, 38, 1999
|
|
1A5E
 
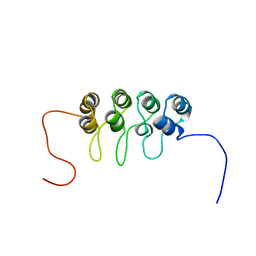 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 18 STRUCTURES | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
1CEH
 
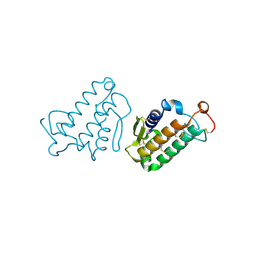 | | STRUCTURE AND FUNCTION OF THE CATALYTIC SITE MUTANT ASP99ASN OF PHOSPHOLIPASE A2: ABSENCE OF CONSERVED STRUCTURAL WATER | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Kumar, A, Sekharudu, C, Ramakrishnan, B, Dupureur, C.M, Zhu, H, Tsai, M.-D, Sundaralingam, M. | | Deposit date: | 1994-11-16 | | Release date: | 1995-02-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the catalytic site mutant Asp 99 Asn of phospholipase A2: absence of the conserved structural water.
Protein Sci., 3, 1994
|
|
2BAX
 
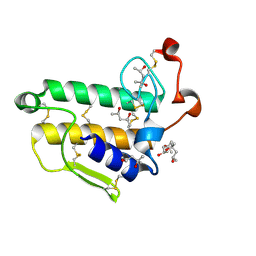 | | Atomic Resolution Structure of the Double Mutant (K53,56M) of Bovine Pancreatic Phospholipase A2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sekar, K, Yogavel, M, Velmurugan, D, Dauter, Z, Dauter, M, Tsai, M.D. | | Deposit date: | 2005-10-15 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution (0.97 A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
4XRH
 
 | |
2B96
 
 | | Third Calcium ion found in an inhibitor bound phospholipase A2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-METHOXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Sekar, K, Velmurugan, D, Yamane, T, Tsai, M.D. | | Deposit date: | 2005-10-11 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Third Calcium ion found in an inhibitor bound phospholipase A2
Acta Crystallogr.,Sect.D, 62, 2006
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
5CWR
 
 | |
5DDY
 
 | | Binary complex of human Polymerase lambda with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA polymerase lambda, MAGNESIUM ION, ... | | Authors: | Liu, M.S, Tsai, M.D. | | Deposit date: | 2015-08-25 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structural Mechanism for the Fidelity Modulation of DNA Polymerase lambda
J.Am.Chem.Soc., 138, 2016
|
|
5DKW
 
 | |
5CA7
 
 | |
1DC2
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 20 STRUCTURES | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR A (P16INK4A) | | Authors: | Byeon, I.-J.L, Li, J, Yuan, C, Tsai, M.-D. | | Deposit date: | 1999-11-04 | | Release date: | 1999-12-23 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: refinement of p16INK4A structure and determination of p15INK4B structure by comparative modeling and NMR data.
Protein Sci., 9, 2000
|
|
