7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6XI3
 
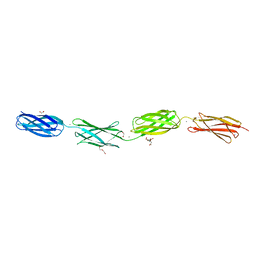 | | Crystal structure of tetra-tandem repeat in extending region of large adhesion protein | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ye, Q, Vance, T.D.R, Davies, P.L. | | Deposit date: | 2020-06-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Essential role of calcium in extending RTX adhesins to their target.
J Struct Biol X, 4, 2020
|
|
6R62
 
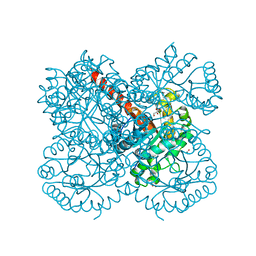 | | Crystal structure of a class II pyruvate aldolase from Sphingomonas wittichii RW1 in complex with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, ACETATE ION, BROMIDE ION, ... | | Authors: | Marsden, S.R, Mestrom, L, Hagedoorn, P.L, Bento, I, McMillan, D.G.G, Hanefeld, U. | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | CH-Pi Interactions Promote the Conversion of Hydroxypyruvate in a Class II Pyruvate Aldolase
Adv.Synth.Catal., 2019
|
|
4P7L
 
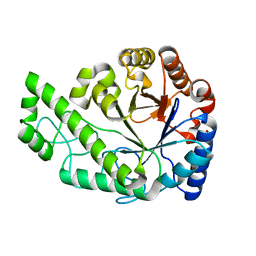 | | Structure of Escherichia coli PgaB C-terminal domain, P212121 crystal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3LZ2
 
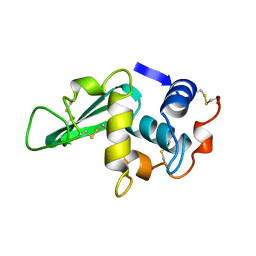 | | STRUCTURE DETERMINATION OF TURKEY EGG WHITE LYSOZYME USING LAUE DIFFRACTION | | Descriptor: | TURKEY EGG WHITE LYSOZYME | | Authors: | Howell, P.L, Almo, S.C, Parsons, M.R, Hajdu, J, Petsko, G.A. | | Deposit date: | 1991-09-13 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination of turkey egg-white lysozyme using Laue diffraction data.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
3VN3
 
 | | Fungal antifreeze protein exerts hyperactivity by constructing an inequable beta-helix | | Descriptor: | 1,2-ETHANEDIOL, Antifreeze protein | | Authors: | Kondo, H, Xiao, N, Hanada, Y, Sugimoto, H, Hoshino, T, Garnham, C.P, Davies, P.L, Tsuda, S. | | Deposit date: | 2011-12-21 | | Release date: | 2012-06-06 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Ice-binding site of snow mold fungus antifreeze protein deviates from structural regularity and high conservation
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6E3Z
 
 | | Structure of Bace-1 in complex with Ligand 8 | | Descriptor: | Beta-secretase 1, N-{3-[(2R,3R)-5-amino-3-methyl-2-(trifluoromethyl)-3,6-dihydro-2H-1,4-oxazin-3-yl]-4-fluorophenyl}-3,5-dichloropyridine-2-carboxamide | | Authors: | Shaffer, P.L. | | Deposit date: | 2018-07-16 | | Release date: | 2019-09-11 | | Last modified: | 2020-03-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and Chemical Development of JNJ-50138803, a Clinical Candidate BACE1 Inhibitor
Acs Symp.Ser., 1307, 2020
|
|
7RZY
 
 | | CryoEM structure of Vibrio cholerae transposon Tn6677 AAA+ ATPase TnsC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Tn6677 Vibrio cholerae transposon TnsC (VchTnsC) | | Authors: | Hoffmann, F.T, Kim, M, Beh, L.Y, Wang, J, Vo, P.L.H, Gelsinger, D.R, Acree, C, Mohabir, J.T, Fernandez, I.S, Sternberg, S.H. | | Deposit date: | 2021-08-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Selective TnsC recruitment enhances the fidelity of RNA-guided transposition.
Nature, 609, 2022
|
|
5WFT
 
 | | PelB 319-436 from Pseudomonas aeruginosa PAO1 | | Descriptor: | PelB | | Authors: | Marmont, L.S, Howell, P.L. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-04 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa.
J. Biol. Chem., 292, 2017
|
|
6LA8
 
 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Lee, P.L, Sharma, D, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
4WCJ
 
 | | Structure of IcaB from Ammonifex degensii | | Descriptor: | CHLORIDE ION, Polysaccharide deacetylase, ZINC ION | | Authors: | Little, D.J, Bamford, N.C, Pokrovskaya, V, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the De-N-acetylation of Poly-beta-1,6-N-acetyl-d-glucosamine in Gram-positive Bacteria.
J.Biol.Chem., 289, 2014
|
|
4WCX
 
 | | Crystal structure of HydG: A maturase of the [FeFe]-hydrogenase | | Descriptor: | ALANINE, Biotin and thiamin synthesis associated, FE (III) ION, ... | | Authors: | Dinis, P.C, Harmer, J.E, Driesener, R.C, Roach, P.L. | | Deposit date: | 2014-09-05 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray crystallographic and EPR spectroscopic analysis of HydG, a maturase in [FeFe]-hydrogenase H-cluster assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2G8E
 
 | | Calpain 1 proteolytic core in complex with SNJ-1715, a cyclic hemiacetal-type inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Calpain-1 catalytic subunit, ... | | Authors: | Cuerrier, D, Moldoveanu, T, Davies, P.L, Campbell, R.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Calpain Inhibition by alpha-Ketoamide and Cyclic Hemiacetal Inhibitors Revealed by X-ray Crystallography
Biochemistry, 45, 2006
|
|
2GBL
 
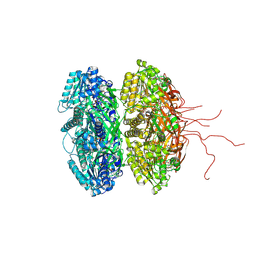 | | Crystal Structure of Full Length Circadian Clock Protein KaiC with Phosphorylation Sites | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Williams, D.R, Pattanayek, S, Xu, Y, Mori, T, Johnson, C.H, Stewart, P.L, Egli, M. | | Deposit date: | 2006-03-10 | | Release date: | 2007-01-23 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of KaiA-KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods.
Embo J., 25, 2006
|
|
4GOL
 
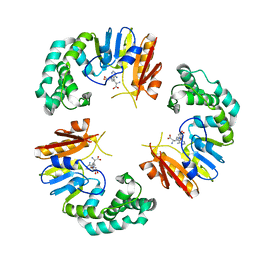 | |
4GOO
 
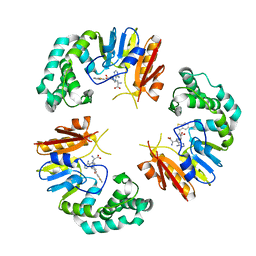 | |
3K90
 
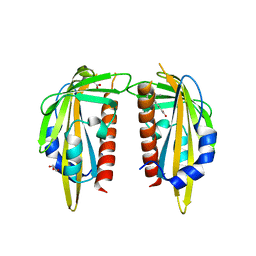 | | The Abscisic acid receptor PYR1 in complex with Abscisic Acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ACETIC ACID, GLYCEROL, ... | | Authors: | Dupeux, F.D, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | Deposit date: | 2009-10-15 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The abscisic acid receptor PYR1 in complex with abscisic acid.
Nature, 462, 2009
|
|
4U0P
 
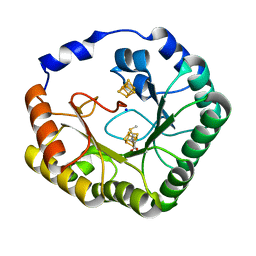 | | The Crystal Structure of Lipoyl Synthase in Complex with S-Adenosyl Homocysteine | | Descriptor: | IRON/SULFUR CLUSTER, Lipoyl synthase 2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Harmer, J.E, Hiscox, M.J, Sandy, J, Dinis, P.C, Roach, P.L. | | Deposit date: | 2014-07-13 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions.
Biochem.J., 464, 2014
|
|
4U0O
 
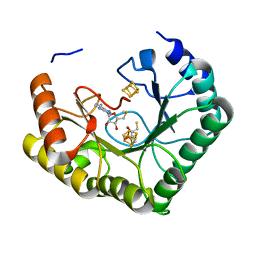 | | Crystal structure of Thermosynechococcus elongatus Lipoyl Synthase 2 complexed with MTA and DTT | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5'-DEOXY-5'-METHYLTHIOADENOSINE, IRON/SULFUR CLUSTER, ... | | Authors: | Harmer, J.E, Hiscox, M.J, Dinis, P.C, Sandy, J, Roach, P.L. | | Deposit date: | 2014-07-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions.
Biochem.J., 464, 2014
|
|
3LGS
 
 | |
7N4N
 
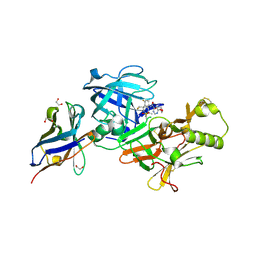 | | BACE-2 in complex with ligand 36 | | Descriptor: | 1,2-ETHANEDIOL, Beta-secretase 2, N-{3-[(2S,5R)-6-amino-2-(fluoromethyl)-5-(methanesulfonyl)-5-methyl-2,3,4,5-tetrahydropyridin-2-yl]-4-fluorophenyl}-6-methoxypyrimidine-4-carboxamide, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2021-06-04 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | JNJ-67569762, A 2-Aminotetrahydropyridine-Based Selective BACE1 Inhibitor Targeting the S3 Pocket: From Discovery to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7N66
 
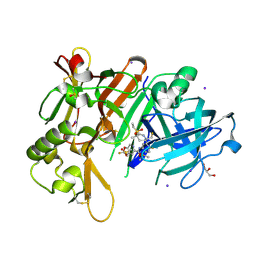 | | BACE-1 in complex with ligand 12 | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2021-06-07 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | JNJ-67569762, A 2-Aminotetrahydropyridine-Based Selective BACE1 Inhibitor Targeting the S3 Pocket: From Discovery to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
6SHT
 
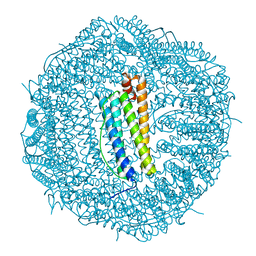 | | Molecular structure of mouse apoferritin resolved at 2.7 Angstroms with the Glacios cryo-microscope | | Descriptor: | FE (III) ION, Ferritin heavy chain, MAGNESIUM ION | | Authors: | Hamdi, F, Tueting, C, Semchonok, D, Kyrilis, F, Meister, A, Skalidis, I, Schmidt, L, Parthier, C, Stubbs, M.T, Kastritis, P.L. | | Deposit date: | 2019-08-08 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | 2.7 angstrom cryo-EM structure of vitrified M. musculus H-chain apoferritin from a compact 200 keV cryo-microscope.
Plos One, 15, 2020
|
|
3DFJ
 
 | | Crystal structure of human Prostasin | | Descriptor: | CHLORIDE ION, Prostasin | | Authors: | Su, H.P, Rickert, K.W, Darke, P.L, Munshi, S.K. | | Deposit date: | 2008-06-12 | | Release date: | 2008-10-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of human prostasin, a target for the regulation of hypertension.
J.Biol.Chem., 283, 2008
|
|
3DF9
 
 | |
