4V7E
 
 | | Model of the small subunit RNA based on a 5.5 A cryo-EM map of Triticum aestivum translating 80S ribosome | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S10E, ... | | Authors: | Barrio-Garcia, C, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion.
Nature, 506, 2014
|
|
1VIT
 
 | | THROMBIN:HIRUDIN 51-65 COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA THROMBIN, ... | | Authors: | Vitali, J, Edwards, B.F.P. | | Deposit date: | 1996-01-31 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a bovine thrombin-hirudin51-65 complex determined by a combination of molecular replacement and graphics. Incorporation of known structural information in molecular replacement.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4V6U
 
 | | Promiscuous behavior of proteins in archaeal ribosomes revealed by cryo-EM: implications for evolution of eukaryotic ribosomes | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10P, ... | | Authors: | Armache, J.-P, Anger, A.M, Marquez, V, Frankenberg, S, Froehlich, T, Villa, E, Berninghausen, O, Thomm, M, Arnold, G.J, Beckmann, R, Wilson, D.N. | | Deposit date: | 2012-08-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Promiscuous behaviour of archaeal ribosomal proteins: Implications for eukaryotic ribosome evolution.
Nucleic Acids Res., 41, 2013
|
|
8S5N
 
 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 12 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
1VRP
 
 | | The 2.1 Structure of T. californica Creatine Kinase Complexed with the Transition-State Analogue Complex, ADP-Mg 2+ /NO3-/Creatine | | Descriptor: | (DIAMINOMETHYL-METHYL-AMINO)-ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, Creatine Kinase, ... | | Authors: | Lahiri, S.D, Wang, P.F, Babbitt, P.C, McLeish, M.J, Kenyon, G.L, Allen, K.N. | | Deposit date: | 2005-04-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A Structure of Torpedo californica Creatine Kinase Complexed with the ADP-Mg(2+)-NO3(-)-Creatine Transition-State Analogue Complex
Biochemistry, 41, 2002
|
|
6Z6B
 
 | | Structure of full-length La Crosse virus L protein (polymerase) | | Descriptor: | RNA (5'-R(*GP*CP*UP*AP*CP*UP*AP*A)-3'), RNA (5'-R(P*AP*GP*UP*AP*GP*UP*GP*UP*GP*C)-3'), RNA (5'-R(P*UP*UP*AP*GP*UP*AP*GP*UP*AP*CP*AP*CP*UP*AP*CP*U)-3'), ... | | Authors: | Cusack, S, Gerlach, P, Reguera, J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.961 Å) | | Cite: | Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes.
Nat Commun, 11, 2020
|
|
3H5Y
 
 | | Norovirus polymerase+primer/template+CTP complex at 6 mM MnCl2 | | Descriptor: | 5'-R(*UP*GP*CP*CP*CP*GP*GP*G)-3', 5'-R(P*UP*GP*CP*CP*CP*GP*GP*GP*C)-3', CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Zamyatkin, D.F, Parra, F, Machin, A, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site.
J.Mol.Biol., 390, 2009
|
|
8S52
 
 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 10 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
1MAA
 
 | | MOUSE ACETYLCHOLINESTERASE CATALYTIC DOMAIN, GLYCOSYLATED PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Taylor, P, Bougis, P.E, Marchot, P. | | Deposit date: | 1998-11-04 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of mouse acetylcholinesterase. A peripheral site-occluding loop in a tetrameric assembly.
J.Biol.Chem., 274, 1999
|
|
3H5X
 
 | | Crystal Structure of 2'-amino-2'-deoxy-cytidine-5'-triphosphate bound to Norovirus GII RNA polymerase | | Descriptor: | 2'-amino-2'-deoxycytidine 5'-(tetrahydrogen triphosphate), 5'-R(*UP*GP*CP*CP*CP*GP*GP*G)-3', 5'-R(P*UP*GP*CP*CP*CP*GP*GP*GP*C)-3', ... | | Authors: | Zamyatkin, D.F, Parra, F, Machin, A, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site.
J.Mol.Biol., 390, 2009
|
|
1VYP
 
 | | Structure of pentaerythritol tetranitrate reductase W102F mutant and complexed with picric acid | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
3UR2
 
 | | Crystal Structure of PTE mutant H254G/H257W/L303T/K185R/I274N/A80V | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, IMIDAZOLE, ... | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
6Z7B
 
 | | Variant Surface Glycoprotein VSGsur bound to suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Variant surface glycoprotein Sur, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E, Jeffrey, P. | | Deposit date: | 2020-05-30 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
3UY9
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-06 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
1MV8
 
 | | 1.55 A crystal structure of a ternary complex of GDP-mannose dehydrogenase from Psuedomonas aeruginosa | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, GDP-mannose 6-dehydrogenase, ... | | Authors: | Snook, C.F, Tipton, P.A, Beamer, L.J. | | Deposit date: | 2002-09-24 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of GDP-mannose
dehydrogenase: A key enzyme in alginate
biosynthesis of P. aeruginosa
Biochemistry, 42, 2003
|
|
7M6F
 
 | |
4OPB
 
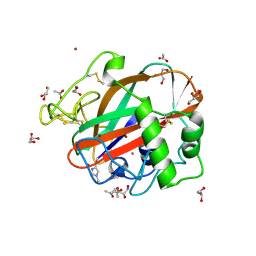 | | AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Lo Leggio, L, Frandsen, K.H, Davies, G.J, Dupree, P, Walton, P, Henrissat, B, Stringer, M, Tovborg, M, De Maria, L, Johansen, K.S. | | Deposit date: | 2014-02-05 | | Release date: | 2015-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase.
Nat Commun, 6, 2015
|
|
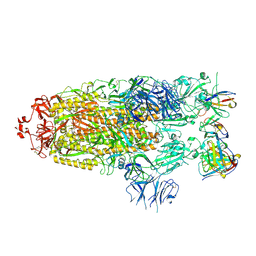
7M6G
 
 | |
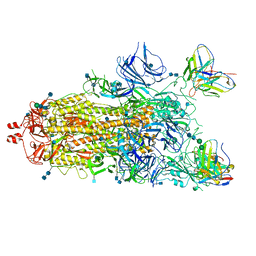
7M6I
 
 | |
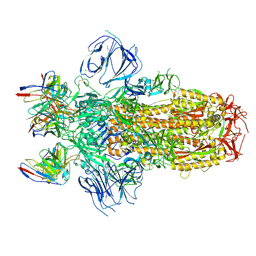
7M6E
 
 | |
7M6H
 
 | |
7M6D
 
 | |
9F3D
 
 | |
4US7
 
 | | Sulfur SAD Phased Structure of a Type IV Pilus Protein from Shewanella oneidensis | | Descriptor: | PILD PROCESSED PROTEIN, SODIUM ION, SULFATE ION | | Authors: | Gorgel, M, Boeggild, A, Ulstrup, J.J, Mueller, U, Weiss, M, Nissen, P, Boesen, T. | | Deposit date: | 2014-07-03 | | Release date: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | High-Resolution Structure of a Type Iv Pilin from the Metal- Reducing Bacterium Shewanella Oneidensis.
Bmc Struct.Biol., 15, 2015
|
|
4NYX
 
 | | Crystal Structure of the Bromodomain of human CREBBP in complex with a dihydroquinoxalinone ligand | | Descriptor: | (3R)-N-[3-(7-methoxy-3,4-dihydroquinolin-1(2H)-yl)propyl]-3-methyl-2-oxo-1,2,3,4-tetrahydroquinoxaline-5-carboxamide, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Rooney, T.P.C, Fedorov, O, Martin, S, Monteiro, O.P, Conway, S.J, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of the Bromodomain of human CREBBP in complex with a dihydroquinoxalinone ligand
TO BE PUBLISHED
|
|
