6G1N
 
 | |
5U1F
 
 | | Initial contact of HIV-1 Env with CD4: Cryo-EM structure of BG505 DS-SOSIP trimer in complex with CD4 and antibody PGT145 | | Descriptor: | BG505 DS-SOSIP gp120, BG505 SOSIP gp41, PGT145 heavy chain, ... | | Authors: | Acharya, P, Kwong, P.D, Potter, C.S, Carragher, B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-02-22 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6W98
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, CALCIUM ION, ... | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-22 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6GA1
 
 | | Bacteriorhodopsin, dark state, cell 1 | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
4PC0
 
 | | Structure of the human RbAp48-MTA1(670-711) complex | | Descriptor: | CALCIUM ION, GLYCEROL, Histone-binding protein RBBP4, ... | | Authors: | Alqarni, S.S.M, Silva, A.P.G, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
6GAB
 
 | | BACTERIORHODOPSIN, 460 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
5DZJ
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T206 in conformation A | | Descriptor: | (2~{R},3~{R},4~{R})-4-methyl-3-(2-oxidanylidene-2-propoxy-ethyl)sulfanyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, L,D-transpeptidase 2 | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5E33
 
 | | Structure of human DPP3 in complex with met-enkephalin | | Descriptor: | Dipeptidyl peptidase 3, MAGNESIUM ION, Met-enkephalin, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-01 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.837 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
6BU9
 
 | | Drosophila Dicer-2 bound to blunt dsRNA | | Descriptor: | Dicer-2, isoform A, RNA (5'-R(*AP*CP*UP*AP*CP*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*C)-3'), ... | | Authors: | Shen, P.S, Sinha, N.K, Bass, B.L. | | Deposit date: | 2017-12-09 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Dicer uses distinct modules for recognizing dsRNA termini.
Science, 359, 2018
|
|
5MJ3
 
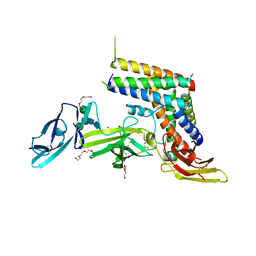 | | INTERLEUKIN-23 COMPLEX WITH AN ANTAGONISTIC ALPHABODY, CRYSTAL FORM 1 | | Descriptor: | ALPHABODY MA12, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | Authors: | Desmet, J, Verstraete, K, Bloch, Y, Lorent, E, Wen, Y, Devreese, B, Vandenbroucke, K, Loverix, S, Hettmann, T, Deroo, S, Somers, K, Hendrikx, P, Lasters, I, Savvides, S.N. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Basis Of Il-23 Antagonism By An Alphabody Protein Scaffold.
Nat Commun, 5, 2014
|
|
7QXM
 
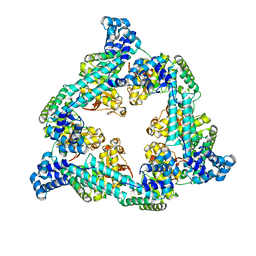 | |
5MJU
 
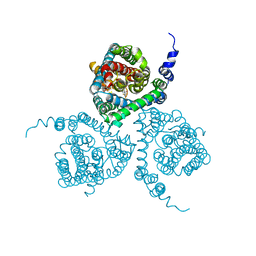 | | Structure of the thermostabilized EAAT1 cryst mutant in complex with the competititve inhibitor TFB-TBOA and the allosteric inhibitor UCPH101 | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[[3-[[4-(trifluoromethyl)phenyl]carbonylamino]phenyl]methoxy]butanedioic acid, 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1 | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-12-01 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
2J2C
 
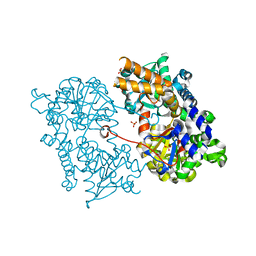 | | Crystal structure of Human Cytosolic 5'-Nucleotidase II (NT5C2, cN-II) | | Descriptor: | CYTOSOLIC PURINE 5'-NUCLEOTIDASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wallden, K, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Karlberg, T, Kotenyova, T, Loppnau, P, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2006-08-16 | | Release date: | 2006-09-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Human Cytosolic 5'- Nucleotidase II: Insights Into Allosteric Regulation and Substrate Recognition
J.Biol.Chem., 282, 2007
|
|
3KBF
 
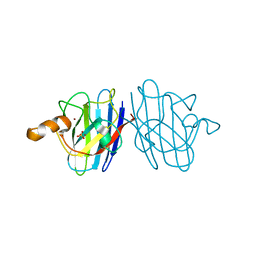 | | C. elegans Cu,Zn Superoxide Dismutase | | Descriptor: | COPPER (II) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Pakhomova, O.N, Taylor, A.B, Schuermann, J.P, Culotta, V.L, Hart, P.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Crystal Structure of C. elegans Cu,Zn Superoxide Dismutase
To be Published
|
|
5HSV
 
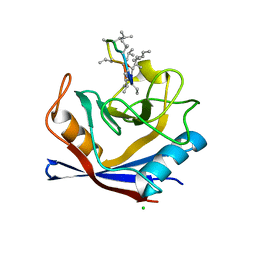 | | X-Ray structure of a CypA-Alisporivir complex at 1.5 angstrom resolution | | Descriptor: | Alisporivir, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Dujardin, M, Bouckaert, J, Rucktooa, P, Hanoulle, X. | | Deposit date: | 2016-01-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of alisporivir in complex with cyclophilin A at 1.5 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7QQ5
 
 | |
7Q6K
 
 | |
5E8E
 
 | | Crystal structure of thrombin bound to an exosite 1-specific IgA Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Baglin, T.P, Langdown, J, Frasson, R, Huntington, J.A. | | Deposit date: | 2015-10-14 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and characterization of an antibody directed against exosite I of thrombin.
J.Thromb.Haemost., 14, 2016
|
|
3ZVY
 
 | | PHD finger of human UHRF1 in complex with unmodified histone H3 N- terminal tail | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE UHRF1, ... | | Authors: | Lallous, N, Birck, C, Mc Ewen, A.G, Legrand, P, Samama, J.P. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Phd Finger of Human Uhrf1 Reveals a New Subgroup of Unmethylated Histone H3 Tail Readers.
Plos One, 6, 2011
|
|
4BEW
 
 | | SERCA bound to phosphate analogue | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Drachmann, N.D, Mattle, D, Laursen, M, Bublitz, M, Olesen, C, Moeller, J.V, Nissen, P, Morth, J.P. | | Deposit date: | 2013-03-12 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Serca Bound to Phosphate Analogue
To be Published
|
|
4PVA
 
 | | Crystal structure of GH62 hydrolase from thermophilic fungus Scytalidium thermophilum | | Descriptor: | GH62 hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Nocek, B, Kaur, A.P, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2014-03-15 | | Release date: | 2014-11-19 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Functional and structural diversity in GH62 alpha-L-arabinofuranosidases from the thermophilic fungus Scytalidium thermophilum.
Microb Biotechnol, 8, 2015
|
|
1P5D
 
 | |
4A5G
 
 | | Raphanus sativus anionic peroxidase. | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jimenez-Arroyo, N, Valderrama, B, Gil-Rodriguez, P, Rojas-Trejo, S.P, Rudino-Pinera, E. | | Deposit date: | 2011-10-25 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic Structure of the Raphanus Sativus Anionic Peroxidase
To be Published
|
|
7QWZ
 
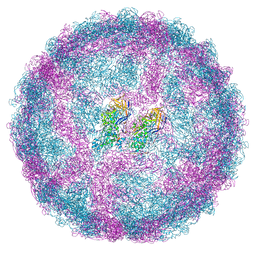 | | Full capsid of Saccharomyces cerevisiae virus L-BCLa | | Descriptor: | Major capsid protein | | Authors: | Grybchuk, D, Prochazkova, M, Fuzik, T, Konovalovas, A, Serva, S, Yurchenko, V, Plevka, P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of L-BC virus and its open particle provide insight into Totivirus capsid assembly.
Commun Biol, 5, 2022
|
|
2YLQ
 
 | | TARGETING THE BINDING FUNCTION 3 SITE OF THE ANDROGEN RECEPTOR THROUGH IN SILICO MOLECULAR MODELING | | Descriptor: | 3-[1-[2-(4-METHYLPHENOXY)ETHYL]BENZIMIDAZOL-2-YL]SULFANYLPROPANOIC ACID, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | Lack, N.A, Axerio, P, Tavassoli, P, Kuchenbecker, K, Han, F.Q, Chan, K.H, Feau, C, LeBlanc, E, Tomlinson, E, Guy, R.K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2011-06-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Binding Function 3 (Bf3) Site of the Human Androgen Receptor Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
