6T5Y
 
 | | Crystal structure of AmpC from E.coli with Zidebactam (WCK 5107) | | Descriptor: | (2S,5R)-1-formyl-N'-[(3R)-piperidine-3-carbonyl]-5-[(sulfooxy)amino]piperidine-2-carbohydrazide, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Lang, P.A, Leissing, T.M, Schofield, C.J, Brem, J. | | Deposit date: | 2019-10-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Investigations of the Inhibition of Escherichia coli AmpC beta-Lactamase by Diazabicyclooctanes.
Antimicrob.Agents Chemother., 65, 2021
|
|
8W1C
 
 | | Cryo-EM structure of BTV pre-subcore | | Descriptor: | Core protein VP3, VP6 | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
6HMX
 
 | |
7UK0
 
 | | [T:Au+:T--pH 11 w/ HgCl2] Metal-mediated DNA base pair in tensegrity triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.36 Å) | | Cite: | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
6F9O
 
 | | Crystal structure of cold-adapted haloalkane dehalogenase DpcA from Psychrobacter cryohalolentis K5 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Haloalkane dehalogenase, ... | | Authors: | Tratsiak, K, Prudnikova, T, Drienovska, I, Damborsky, J, Brynda, J, Pachl, P, Kuty, M, Chaloupkova, R, Kuta Smatanova, I, Rezacova, P. | | Deposit date: | 2017-12-15 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure of the cold-adapted haloalkane dehalogenase DpcA from Psychrobacter cryohalolentis K5.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8G1Q
 
 | | Co-crystal structure of Compound 1 in complex with the bromodomain of human SMARCA4 and pVHL:ElonginC:ElonginB | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongin-B, Elongin-C, ... | | Authors: | Ghimire Rijal, S, Wurz, R.P, Vaish, A. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Affinity and cooperativity modulate ternary complex formation to drive targeted protein degradation.
Nat Commun, 14, 2023
|
|
3KBS
 
 | |
8W1S
 
 | | Cryo-EM structure of BTV pre-core | | Descriptor: | Core protein VP3, RNA-directed RNA polymerase | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8BP8
 
 | | SPA of Trypsin untreated Rotavirus TLP spike | | Descriptor: | CALCIUM ION, Inner capsid protein VP2, Intermediate capsid protein VP6, ... | | Authors: | Shah, P.N.M, Stuart, D.I. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography.
Cell Host Microbe, 31, 2023
|
|
3KBV
 
 | |
3KBM
 
 | |
6YE0
 
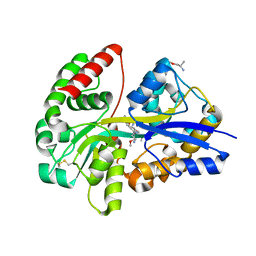 | | E.coli's Putrescine receptor PotF complexed with Putrescine | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{R})-1-[(2~{R})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-amine, (2~{S})-1-(2-methoxyethoxy)propan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-23 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
8GUD
 
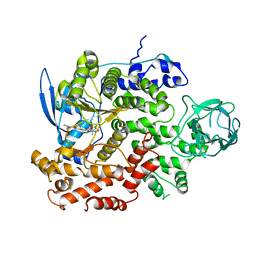 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant E545K in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GUB
 
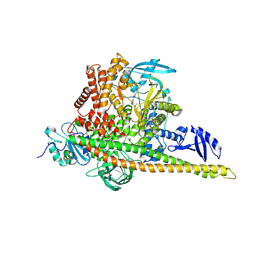 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant H1047R in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6FBR
 
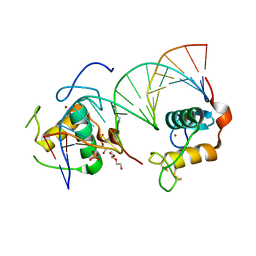 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human MEp DR1 Response Element, pH 4.2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*TP*GP*GP*GP*TP*CP*AP*AP*AP*GP*TP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*AP*CP*TP*TP*TP*GP*AP*CP*CP*CP*AP*G)-3'), ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2017-12-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modulation of RXR-DNA complex assembly by DNA context.
Mol. Cell. Endocrinol., 481, 2019
|
|
8GRH
 
 | | Crystal structure of a constitutively active mutant of the alpha gamma heterodimer of human IDH3 in complex with CIT | | Descriptor: | CITRIC ACID, Human IDH3 alpha subunit, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
1AP1
 
 | |
5YL5
 
 | | Crystal structure of dodecameric Dehydroquinate dehydratase from Acinetobacter baumannii at 1.9A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinate dehydratase, GLYCEROL, ... | | Authors: | Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dodecameric Dehydroquinate dehydratase from Acinetobacter baumannii at 1.9A resolution
To Be Published
|
|
8PN9
 
 | | Structure of human oligosaccharyltransferase OST-A complex bound to NGI-1 | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-[(1~{S},2~{R},3~{R},4~{R},5'~{S},6~{S},7~{R},8~{S},9~{R},12~{R},13~{R},15~{S},16~{S},18~{R})-5',7,9,13-tetramethyl-3,15-bis(oxidanyl)spiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icosane-6,2'-oxane]-16-yl]oxy-oxane-3,4,5-triol, (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphadocosan-1-aminium, ... | | Authors: | Ramirez, A.S, Kowal, J, Locher, K.P. | | Deposit date: | 2023-06-30 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Positive selection CRISPR screens reveal a druggable pocket in an oligosaccharyltransferase required for inflammatory signaling to NF-kappa B.
Cell, 187, 2024
|
|
3ZLO
 
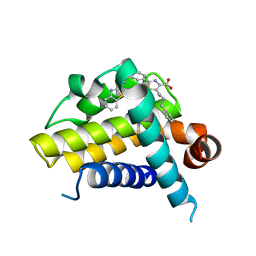 | | Crystal structure of BCL-XL in complex with inhibitor (Compound 6) | | Descriptor: | 2-[(8E)-8-(1,3-benzothiazol-2-ylhydrazinylidene)-6,7-dihydro-5H-naphthalen-2-yl]-5-(4-phenylbutyl)-1,3-thiazole-4-carboxylic acid, BCL-2-LIKE PROTEIN 1 | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-02-04 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
6VMS
 
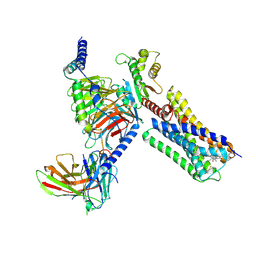 | | Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane | | Descriptor: | Endolysin,D(2) dopamine receptor,D(2) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yin, J, Chen, K.M, Clark, M.J, Hijazi, M, Kumari, P, Bai, X, Sunahara, R.K, Barth, P, Rosenbaum, D.M. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-17 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane.
Nature, 584, 2020
|
|
6VXZ
 
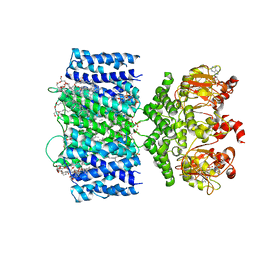 | | SthK P300A cyclic nucleotide-gated potassium channel in the closed state, in complex with cAMP | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, SthK | | Authors: | Schmidpeter, P.A.M, Rheinberger, J, Nimigean, C.M. | | Deposit date: | 2020-02-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Prolyl isomerization controls activation kinetics of a cyclic nucleotide-gated ion channel.
Nat Commun, 11, 2020
|
|
8GUA
 
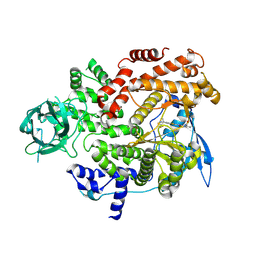 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant E542K in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2VQW
 
 | | Structure of inhibitor-free HDAC4 catalytic domain (with gain-of- function mutation His332Tyr) | | Descriptor: | HISTONE DEACETYLASE 4, POTASSIUM ION, ZINC ION | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-19 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Structural Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQM
 
 | | Structure of HDAC4 catalytic domain bound to a hydroxamic acid inhbitor | | Descriptor: | HISTONE DEACETYLASE 4, N-hydroxy-5-[(3-phenyl-5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)carbonyl]thiophene-2-carboxamide, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
