9EP4
 
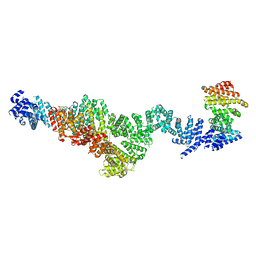 | | Structure of Integrator subcomplex INTS5/8/15 | | Descriptor: | Integrator complex subunit 15, Integrator complex subunit 5, Integrator complex subunit 8 | | Authors: | Razew, M, Galej, W.P. | | Deposit date: | 2024-03-17 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of the Integrator complex assembly and association with transcription factors.
Mol.Cell, 2024
|
|
9EY3
 
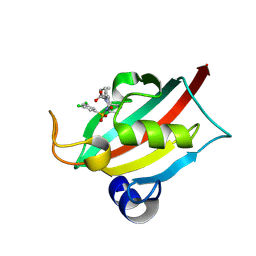 | | The FK1 domain of FKBP51 in complex with (3S,11S,11aS)-12-((3,5-dichlorophenyl)sulfonyl)-5-oxo-11-vinyldecahydro-1H-6,10-epiminopyrrolo[1,2-a]azonine-3-carboxylic acid | | Descriptor: | (1~{S},4~{S},7~{S},8~{S},9~{R})-13-[3,5-bis(chloranyl)phenyl]sulfonyl-8-ethenyl-2-oxidanylidene-3,13-diazatricyclo[7.3.1.0^{3,7}]tridecane-4-carboxylic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Krajczy, P, Hausch, F. | | Deposit date: | 2024-04-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure-Based Design of Ultrapotent Tricyclic Ligands for FK506-Binding proteins.
Chemistry, 2024
|
|
9FA4
 
 | |
8YZK
 
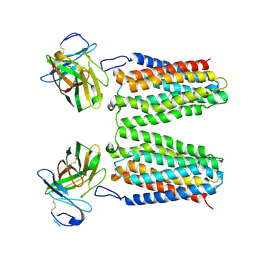 | | Orphan receptor GPRC5D in complex with scFv150-18 | | Descriptor: | Soluble cytochrome b562,G-protein coupled receptor family C group 5 member D, scFv | | Authors: | Yan, P, Lin, X, Xu, F. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The binding mechanism of an anti-multiple myeloma antibody to the human GPRC5D homodimer.
Nat Commun, 15, 2024
|
|
9FHP
 
 | |
8XH6
 
 | | Structure of EBV LMP1 dimer | | Descriptor: | Latent membrane protein 1 | | Authors: | Gao, P, Huang, J.F. | | Deposit date: | 2023-12-17 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Assembly and activation of EBV latent membrane protein 1
To Be Published
|
|
8Z9A
 
 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum bound with geranyl acetate | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
7QB2
 
 | | Pim1 in complex with (E)-4-((6-amino-1-methyl-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(E)-(6-azanyl-1-methyl-2-oxidanylidene-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
7XKW
 
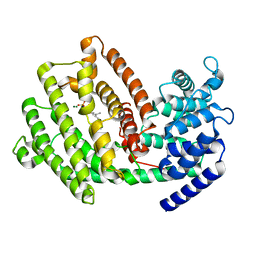 | | The 3D strcuture of (-)-cyperene synthase with substrate analogue FSPP | | Descriptor: | (-)-cyperene synthase, MAGNESIUM ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE | | Authors: | Yu, S.S, Zhu, P, Liu, Y.B, Ma, S.G, Ye, D, Shao, Y.Z, Li, W.R, Cui, Z.J. | | Deposit date: | 2022-04-20 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Characterization and Engineering of Two Highly Paralogous Sesquiterpene Synthases Reveal a Regioselective Reprotonation Switch.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
9BQV
 
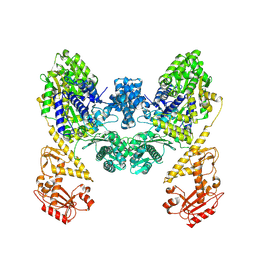 | | DdmD dimer apoprotein | | Descriptor: | Helicase/UvrB N-terminal domain-containing protein | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
9BD3
 
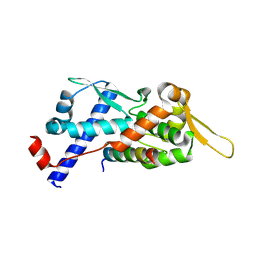 | | Structure of the MAGEA4 MHD-RAD18 R6BD Complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Melanoma antigen A 4 | | Authors: | Forker, K, Zhou, P. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of MAGEA4 MHD-RAD18 R6BD reveals a flipped binding mode compared to AlphaFold2 prediction.
Embo J., 2024
|
|
3GKE
 
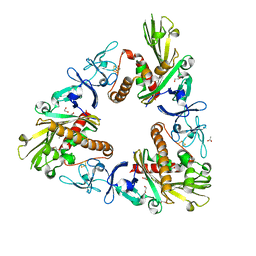 | | Crystal Structure of Dicamba Monooxygenase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DdmC, ... | | Authors: | Wilson, M.A, Dumitru, R, Jiang, W.Z, Weeks, D.P. | | Deposit date: | 2009-03-10 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dicamba monooxygenase: a Rieske nonheme oxygenase that catalyzes oxidative demethylation.
J.Mol.Biol., 392, 2009
|
|
5VII
 
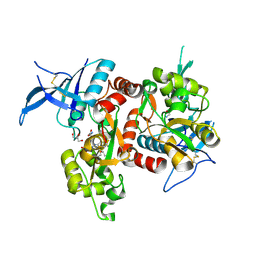 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-(3-fluoropropyl)phenyl-ACEPC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(2R)-2-amino-2-carboxyethyl]-1-[4-(3-fluoropropyl)phenyl]-1H-pyrazole-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6KCK
 
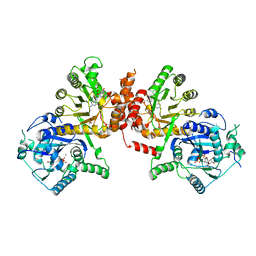 | | Crystal structure of Plasmodium falciparum HPPK-DHPS wild type with pterin and p-hydroxybenzoate | | Descriptor: | 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ACETATE ION, CALCIUM ION, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
9BBE
 
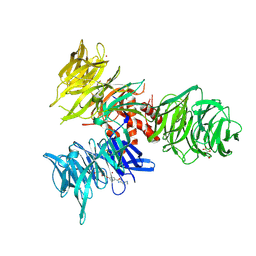 | | Co-crystal structure of human DDB1 bound to fragment UB028668 | | Descriptor: | 5-(4-methoxyphenyl)-3-[(3S)-pyrrolidin-3-yl]-1,2,4-oxadiazole, DNA damage-binding protein 1, L(+)-TARTARIC ACID, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028668
To be published
|
|
7QFM
 
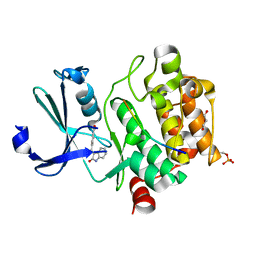 | | Pim1 in complex with (E)-4-((2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
8ZMM
 
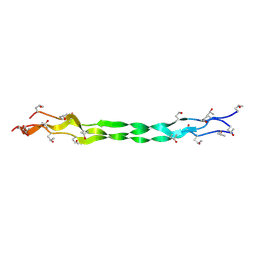 | | Structure of a triple-helix region of human Collagen type IV from Trautec | | Descriptor: | collagen type IV | | Authors: | Chu, Y, Zhai, Y, Fan, X, Fu, S, Li, J, Wu, X, Cai, H, Wang, X, Li, D, Feng, P, Cao, K, Qian, S. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a triple-helix region of human Collagen type IV from Trautec
To Be Published
|
|
5VF1
 
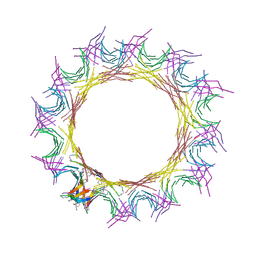 | |
5VGB
 
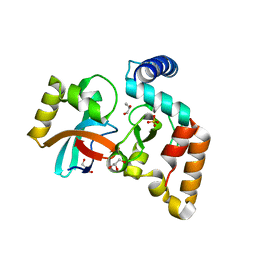 | | Crystal structure of NmeCas9 HNH domain bound to anti-CRISPR AcrIIC1 | | Descriptor: | Anti-CRISPR protein (AcrIIC1), CRISPR-associated endonuclease Cas9, GLYCEROL, ... | | Authors: | Harrington, L.B, Doxzen, K.W, Ma, E, Knott, G.J, Kranzusch, P.J, Doudna, J.A. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | A Broad-Spectrum Inhibitor of CRISPR-Cas9.
Cell, 170, 2017
|
|
4O1M
 
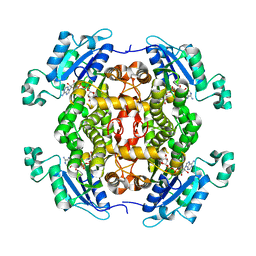 | | Toxoplasma gondii Enoyl acyl carrier protein reductase | | Descriptor: | Enoyl-acyl carrier reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Muench, S.P, Wilkinson, C, Prigge, S.T, Rice, D.W. | | Deposit date: | 2013-12-16 | | Release date: | 2014-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The benzimidazole based drugs show good activity against T. gondii but poor activity against its proposed enoyl reductase enzyme target
Bioorg.Med.Chem.Lett., 24, 2014
|
|
9C61
 
 | | Crystal structure of the human LRRK2 WDR domain in complex with CACHE1193-26 | | Descriptor: | (4R)-4-[4-(5-fluoro-1H-indol-3-yl)piperidine-1-carbonyl]piperidin-2-one, Non-specific serine/threonine protein kinase, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Kutera, M, Ilyassov, O, Seitova, A, Loppnau, P, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Halabelian, L. | | Deposit date: | 2024-06-07 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the human LRRK2 WDR domain in complex with CACHE1193-26
To be published
|
|
9EN2
 
 | | Crystal structure of the metalloproteinase enhancer PCPE-1 complexed with nanobodies VHH-H4 and VHH-I5 | | Descriptor: | CALCIUM ION, GLYCEROL, Procollagen C-endopeptidase enhancer 1, ... | | Authors: | Lagoutte, P, Gueguen-Chaignon, V, Bourhis, J.-M, Vadon-Le Goff, S. | | Deposit date: | 2024-03-12 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mono- and bi-specific nanobodies targeting the CUB domains of PCPE-1 reduce the proteolytic processing of fibrillar procollagens.
J.Mol.Biol., 2024
|
|
5VKK
 
 | |
4O4J
 
 | | Tubulin-Peloruside A complex | | Descriptor: | CALCIUM ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Bargsten, K, Northcote, P.T, Marsh, M, Altmann, K.H, Miller, J.H, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2013-12-18 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of microtubule stabilization by laulimalide and peloruside A.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
9EME
 
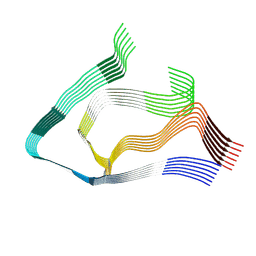 | | AL amyloid fibril from the FOR103 light chain | | Descriptor: | lambda 3 immunoglobulin light chain fragment, residues 2-116 | | Authors: | Pfeiffer, P.B, Karimi-Farsijani, S, Kupfer, N, Schmidt, M, Faendrich, M. | | Deposit date: | 2024-03-08 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Light chain mutations contribute to defining the fibril morphology in systemic AL amyloidosis.
Nat Commun, 15, 2024
|
|
