2M2E
 
 | | Solution NMR structure of the SANT domain of human DNAJC2, Northeast structural genomics consortium target HR8254a | | Descriptor: | DnaJ homolog subfamily C member 2 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Ong, M, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-18 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the SANT domain of human DnaJC2.
To be Published
|
|
5X12
 
 | | Crystal structure of Bacillus subtilis PadR | | Descriptor: | Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
5X11
 
 | | Crystal structure of Bacillus subtilis PadR in complex with operator DNA | | Descriptor: | DNA (28-MER), Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
8ZUG
 
 | |
5X14
 
 | | Crystal structure of Bacillus subtilis PadR in complex with ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, GLYCEROL, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
6LU8
 
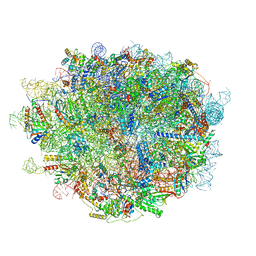 | | Cryo-EM structure of a human pre-60S ribosomal subunit - state A | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-26 | | Release date: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
6LQM
 
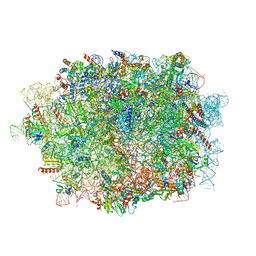 | | Cryo-EM structure of a pre-60S ribosomal subunit - state C | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-14 | | Release date: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
6NZN
 
 | | Dimer-of-dimer amyloid fibril structure of glucagon | | Descriptor: | Glucagon | | Authors: | Gelenter, M.D, Smith, K.J, Liao, S.Y, Mandala, V.S, Dregni, A.J, Lamm, M.S, Tian, Y, Wei, X, Pochan, D.J, Tucker, T.J, Su, Y, Hong, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The peptide hormone glucagon forms amyloid fibrils with two coexisting beta-strand conformations.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5GM8
 
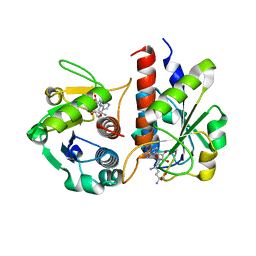 | | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruiginosa | | Descriptor: | SINEFUNGIN, tRNA (cytidine/uridine-2'-O-)-methyltransferase TrmJ | | Authors: | Jaroensuk, J, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Liew, C.W, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa.
Nucleic Acids Res., 44, 2016
|
|
5GMC
 
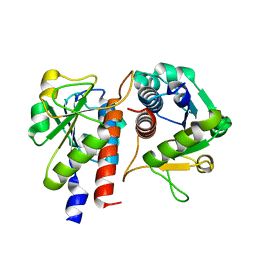 | | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruiginosa | | Descriptor: | tRNA (cytidine/uridine-2'-O-)-methyltransferase TrmJ | | Authors: | Jaroensuk, J, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Liew, C.W, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa.
Nucleic Acids Res., 44, 2016
|
|
5GMB
 
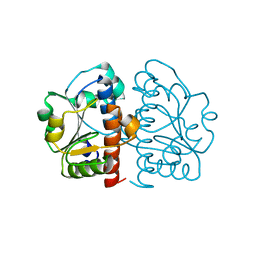 | | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruiginosa | | Descriptor: | tRNA (cytidine/uridine-2'-O-)-methyltransferase TrmJ | | Authors: | Jaroensuk, J, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Liew, C.W, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa.
Nucleic Acids Res., 44, 2016
|
|
8ODQ
 
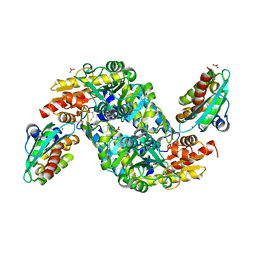 | | SufS-SufU complex from Mycobacterium tuberculosis | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Cysteine desulfurase, NITRATE ION, ... | | Authors: | Elchennawi, I, Carpentier, P, Caux, C, Ponge, M, Ollagnier de Choudens, S. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Characterization of Mycobacterium tuberculosis Zinc SufU-SufS Complex.
Biomolecules, 13, 2023
|
|
7KZ7
 
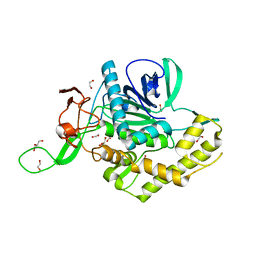 | | Crystals Structure of the Mutated Protease Domain of Botulinum Neurotoxin X (X4130B1). | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin type X, GLYCEROL, ... | | Authors: | Blum, T.R, Liu, H, Packer, M.S, Xiong, X, Lee, P.G, Zhang, S, Richter, M, Minasov, G, Satchell, K.J.F, Dong, M, Liu, D.R, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phage-assisted evolution of botulinum neurotoxin proteases with reprogrammed specificity.
Science, 371, 2021
|
|
9CGX
 
 | | Alzheimer's Disease Seeded 0N3R Tau Fibrils | | Descriptor: | Isoform Fetal-tau of Microtubule-associated protein tau | | Authors: | Duan, P, Dregni, A.J, Xu, H, Changolkar, L, Lee, V.M.-Y, Hong, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Alzheimer's disease seeded tau forms paired helical filaments yet lacks seeding potential.
J.Biol.Chem., 300, 2024
|
|
9CGZ
 
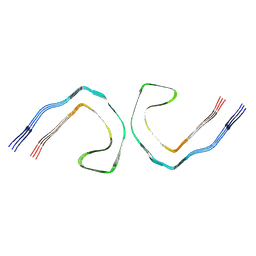 | | Alzheimer's Disease Seeded Mixed 0N4R and 0N3R Tau Fibrils | | Descriptor: | Isoform Fetal-tau of Microtubule-associated protein tau | | Authors: | Duan, P, Dregni, A.J, Xu, H, Changolkar, L, Lee, V.M.-Y, Hong, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Alzheimer's disease seeded tau forms paired helical filaments yet lacks seeding potential.
J.Biol.Chem., 300, 2024
|
|
7T4D
 
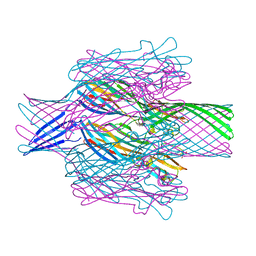 | | Pore structure of pore-forming toxin Epx4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Epx4 | | Authors: | Xiong, X.Z, Dong, M, Yang, P, Abraham, J. | | Deposit date: | 2021-12-09 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Emerging enterococcus pore-forming toxins with MHC/HLA-I as receptors.
Cell, 185, 2022
|
|
7T4E
 
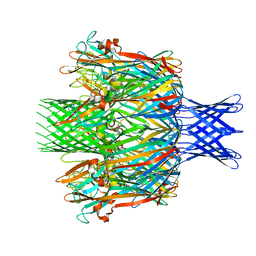 | |
3ZPQ
 
 | | Thermostabilised turkey beta1 adrenergic receptor with 4-(piperazin-1- yl)-1H-indole bound (compound 19) | | Descriptor: | 4-(PIPERAZIN-1-YL)-1H-INDOLE, BETA-1 ADRENERGIC RECEPTOR, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Christopher, J.A, Congreve, M, Dore, A.S, Marshall, F.H, Myszka, D.G, Brown, J, Koglin, M, Tehan, B, Errey, J.C, Tate, C.G, Warne, T. | | Deposit date: | 2013-03-01 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biophysical Fragment Screening of the Beta1-Adrenergic Receptor: Identification of High Affinity Aryl Piperazine Leads Using Structure-Based Drug Design.
J.Med.Chem., 56, 2013
|
|
3ZPR
 
 | | Thermostabilised turkey beta1 adrenergic receptor with 4-methyl-2-(piperazin-1-yl) quinoline bound | | Descriptor: | 4-METHYL-2-(PIPERAZIN-1-YL) QUINOLINE, BETA-1 ADRENERGIC RECEPTOR, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Christopher, J.A, Congreve, M, Dore, A.S, Marshall, F.H, Myszka, D.G, Brown, J, Koglin, M, Tehan, B, Errey, J.C, Tate, C.G, Warne, T. | | Deposit date: | 2013-03-01 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biophysical Fragment Screening of the Beta1-Adrenergic Receptor: Identification of High Affinity Aryl Piperazine Leads Using Structure-Based Drug Design.
J.Med.Chem., 56, 2013
|
|
5HID
 
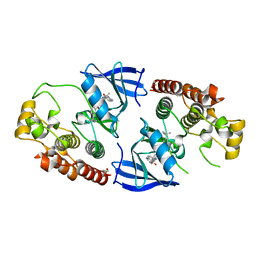 | | BRAF Kinase domain b3aC loop deletion mutant in complex with AZ628 | | Descriptor: | 3-(2-cyanopropan-2-yl)-N-{4-methyl-3-[(3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]phenyl}benzamide, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase B-raf | | Authors: | Whalen, D.M, Foster, S.A, Ozen, A, Wongchenko, M, Yin, J, Schaefer, G, Mayfield, J, Chmielecki, J, Stephens, P, Albacker, L, Yan, Y, Song, K, Hatzivassiliou, G, Eigenbrot, C, Yu, C, Shaw, A.S, Manning, G, Skelton, N.J, Hymowitz, S.G, Malek, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
5HI2
 
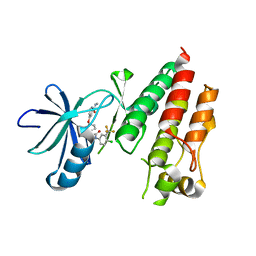 | | BRAF Kinase domain b3aC loop deletion mutant in complex with sorafenib | | Descriptor: | 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, Serine/threonine-protein kinase B-raf | | Authors: | Whalen, D.M, Foster, S.A, Ozen, A, Wongchenko, M, Yin, J, Schaefer, G, Mayfield, J, Chmielecki, J, Stephens, P, Albacker, L, Yan, Y, Song, K, Hatzivassiliou, G, Eigenbrot, C, Yu, C, Shaw, A.S, Manning, G, Skelton, N.J, Hymowitz, S.G, Malek, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
5HIE
 
 | | BRAF Kinase domain b3aC loop deletion mutant in complex with dabrafenib | | Descriptor: | Dabrafenib, Serine/threonine-protein kinase B-raf | | Authors: | Whalen, D.M, Foster, S.A, Ozen, A, Wongchenko, M, Yin, J, Schaefer, G, Mayfield, J, Chmielecki, J, Stephens, P, Albacker, L, Yan, Y, Song, K, Hatzivassiliou, G, Eigenbrot, C, Yu, C, Shaw, A.S, Manning, G, Skelton, N.J, Hymowitz, S.G, Malek, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
4QD2
 
 | | Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex | | Descriptor: | CALCIUM ION, Cadherin-1, Hemagglutinin component HA17, ... | | Authors: | Lee, K, Zhong, X, Gu, S, Kruel, A, Dorner, M.B, Perry, K, Rummel, A, Dong, M, Jin, R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex.
Science, 344, 2014
|
|
5Y8T
 
 | | Crystal structure of Bacillus subtilis PadR in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR.
Nucleic Acids Res., 45, 2017
|
|
7SFQ
 
 | | EmrE S64V Mutant Bound to tetra(4-fluorophenyl)phosphonium at pH 8.0 | | Descriptor: | Multidrug transporter EmrE, tetrakis(4-fluorophenyl)phosphanium | | Authors: | Shcherbakov, A.A, Spreacker, P.J, Dregni, A.J, Henzler-Wildman, K.A, Hong, M. | | Deposit date: | 2021-10-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | High-pH structure of EmrE reveals the mechanism of proton-coupled substrate transport.
Nat Commun, 13, 2022
|
|
