5WXI
 
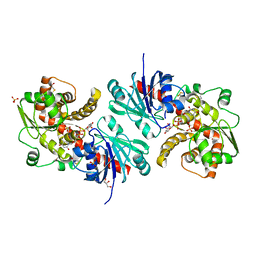 | | EarP bound with dTDP-rhamnose (soaked) | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, BETA-MERCAPTOETHANOL, EarP, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
7X5F
 
 | | Nrf2-MafG heterodimer bound with CsMBE2 | | Descriptor: | Nuclear factor erythroid 2-related factor 2, Synthetic DNA, Transcription factor MafG | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5G
 
 | | Nrf2 (A510Y)-MafG heterodimer bound with CsMBE2 | | Descriptor: | DNA (5'-D(*CP*AP*CP*AP*GP*TP*GP*AP*CP*TP*CP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP*CP*TP*GP*T)-3'), Nuclear factor erythroid 2-related factor 2, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5E
 
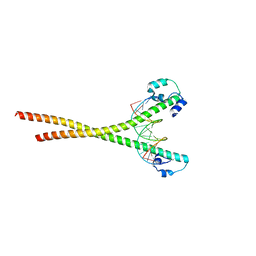 | | Nrf2-MafG heterodimer bound with CsMBE1 | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*T)-3'), HEXAETHYLENE GLYCOL, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
2EJR
 
 | | LSD1-tranylcypromine complex | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL (2R,3S,4S)-5-[7,8-DIMETHYL-2,4-DIOXO-5-(3-PHENYLPROPANOYL)-1,3,4,5-TETRAHYDROBENZO[G]PTERIDIN-10(2H)-YL]-2,3,4-TRIHYDROXYPENTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Sengoku, T, Mimasu, S, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of histone demethylase LSD1 and tranylcypromine at 2.25A
Biochem.Biophys.Res.Commun., 366, 2008
|
|
2DB3
 
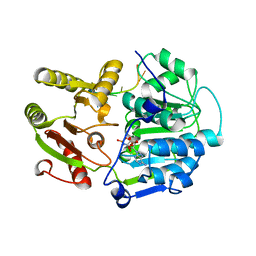 | | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase vasa, MAGNESIUM ION, ... | | Authors: | Sengoku, T, Nureki, O, Nakamura, A, Kobayashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.
Cell(Cambridge,Mass.), 125, 2006
|
|
2DW4
 
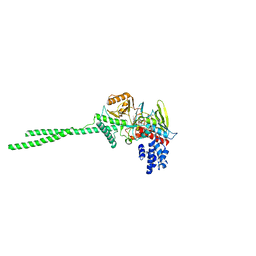 | |
3AVS
 
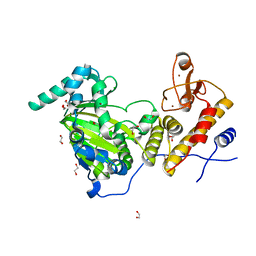 | | Catalytic fragment of UTX/KDM6A bound with N-oxyalylglycine, and Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 6A, ... | | Authors: | Sengoku, T, Yokoyama, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A
Genes Dev., 25, 2011
|
|
3AVR
 
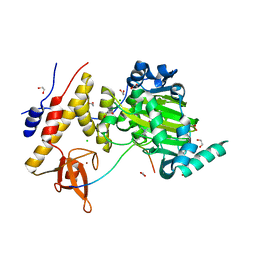 | | Catalytic fragment of UTX/KDM6A bound with histone H3K27me3 peptide, N-oxyalylglycine, and Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone H3, ... | | Authors: | Sengoku, T, Yokoyama, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A
Genes Dev., 25, 2011
|
|
3DMI
 
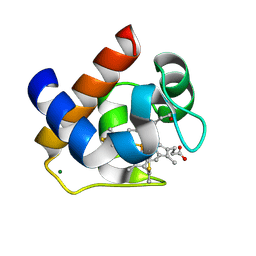 | | Crystallization and Structural Analysis of Cytochrome c6 from the Diatom Phaeodactylum tricornutum at 1.5 A resolution | | Descriptor: | HEME C, MAGNESIUM ION, cytochrome c6 | | Authors: | Akazaki, H, Kawai, F, Hosokawa, M, Hama, T, Hirano, T, Lim, B.-K, Sakurai, N, Hakamata, W, Park, S.-Y, Nishio, T, Oku, T. | | Deposit date: | 2008-07-01 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallization and structural analysis of cytochrome c(6) from the diatom Phaeodactylum tricornutum at 1.5 A resolution.
Biosci.Biotechnol.Biochem., 73, 2009
|
|
1GDV
 
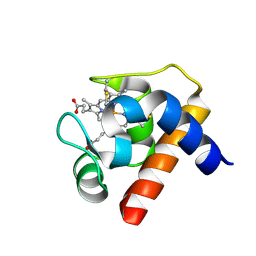 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM RED ALGA PORPHYRA YEZOENSIS AT 1.57 A RESOLUTION | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Yamada, S, Park, S.-Y, Shimizu, H, Shiro, Y, Oku, T. | | Deposit date: | 2000-10-06 | | Release date: | 2001-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of cytochrome c6 from the red alga Porphyra yezoensis at 1. 57 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2DGE
 
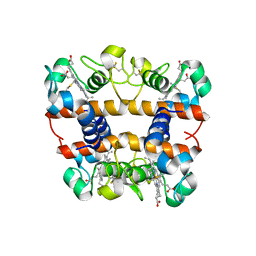 | | Crystal structure of oxidized cytochrome C6A from Arabidopsis thaliana | | Descriptor: | Cytochrome c6, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION | | Authors: | Chida, H, Yokoyama, T, Kawai, F, Nakazawa, A, Akazaki, H, Takayama, Y, Hirano, T, Suruga, K, Satoh, T, Yamada, S, Kawachi, R, Unzai, S, Nishio, T, Park, S.-Y, Oku, T. | | Deposit date: | 2006-03-11 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of oxidized cytochrome c(6A) from Arabidopsis thaliana
Febs Lett., 580, 2006
|
|
3VOV
 
 | | Crystal Structure of ROK Hexokinase from Thermus thermophilus | | Descriptor: | GLYCEROL, Glucokinase, ZINC ION | | Authors: | Nakamura, T, Kashima, Y, Mine, S, Oku, T, Uegaki, K. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and crystal structure of the thermophilic ROK hexokinase from Thermus thermophilus
J.Biosci.Bioeng., 2012
|
|
2ZBO
 
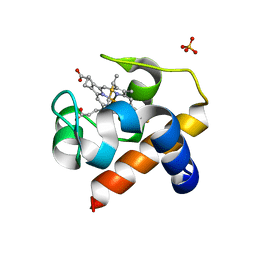 | | Crystal structure of low-redox-potential cytochrom c6 from brown alga Hizikia fusiformis at 1.6 A resolution | | Descriptor: | Cytochrome c6, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Akazaki, H, Kawai, F, Chida, H, Matsumoto, Y, Sirasaki, I, Nakade, H, Hirayama, M, Hosikawa, K, Suruga, K, Satoh, T, Yamada, S, Unzai, S, Hakamata, W, Nishio, T, Park, S.-Y, Oku, T. | | Deposit date: | 2007-10-26 | | Release date: | 2008-09-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cloning, expression and purification of cytochrome c(6) from the brown alga Hizikia fusiformis and complete X-ray diffraction analysis of the structure
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
2ZZS
 
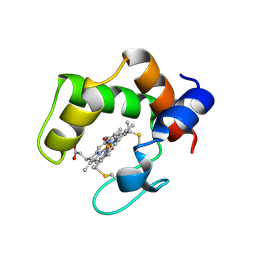 | | Crystal structure of cytochrome c554 from Vibrio parahaemolyticus strain RIMD2210633 | | Descriptor: | Cytochrome c554, GLYCEROL, HEME C | | Authors: | Akazaki, H, Kawai, F, Kumaki, Y, Sekine, K, Hakamata, W, Nishio, T, Park, S.-Y, Oku, T. | | Deposit date: | 2009-02-24 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cytochrome c554 from Vibrio parahaemolyticus strain RIMD2210633
To be Published
|
|
8SKM
 
 | |
1UCQ
 
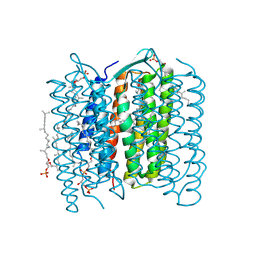 | | Crystal structure of the L intermediate of bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, RETINAL, ... | | Authors: | Kouyama, T, Nishikawa, T, Tokuhisa, T, Okumura, H. | | Deposit date: | 2003-04-17 | | Release date: | 2003-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the L Intermediate of Bacteriorhodopsin: Evidence for Vertical Translocation of a Water Molecule during the Proton Pumping Cycle.
J.Mol.Biol., 335, 2004
|
|
8JE4
 
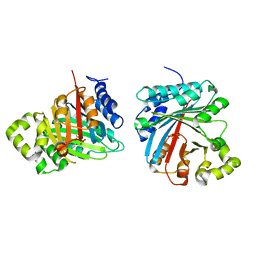 | | Crystal structure of LimF prenyltransferase (H239G/W273T mutant) bound with the thiodiphosphate moiety of farnesyl S-thiolodiphosphate (FSPP) | | Descriptor: | MAGNESIUM ION, TRIHYDROGEN THIODIPHOSPHATE, prenyltransferase, ... | | Authors: | Hamada, K, Oguni, A, Zhang, Y, Satake, M, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Switching Prenyl Donor Specificities of Cyanobactin Prenyltransferases.
J.Am.Chem.Soc., 145, 2023
|
|
8J12
 
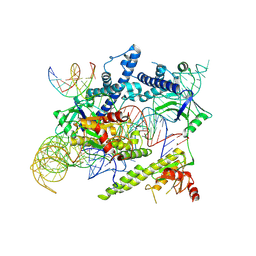 | | Cryo-EM structure of the AsCas12f-sgRNA-target DNA ternary complex | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (247-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-12 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
8J1J
 
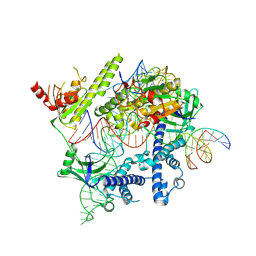 | | Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (118-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
8J3R
 
 | | Cryo-EM structure of the AsCas12f-HKRA-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (37-MER), DNA (38-MER), MAGNESIUM ION, ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-18 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
9FN3
 
 | | Crystal structure of the alkyltransferase ribozyme SAMURI co-crystalized with ProSeDMA | | Descriptor: | MAGNESIUM ION, SAMURI-ProSeDMA, Se-2,6-diaminopurinribosyl-selenohomocysteineamide | | Authors: | Chen, H.A, Okuda, T, Lenz, A.K, Scheitl, C.P.M, Schindelin, H, Hoebartner, C. | | Deposit date: | 2024-06-07 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and catalytic activity of the SAM-utilizing ribozyme SAMURI.
Nat.Chem.Biol., 2025
|
|
4B0A
 
 | | The high-resolution structure of yTBP-yTAF1 identifies conserved and competing interaction surfaces in transcriptional activation | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Anandapadamanaban, M, Andresen, C, Siponen, M, Kokubo, T, Ikura, M, Moche, M, Sunnerhagen, M. | | Deposit date: | 2012-06-29 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | High-Resolution Structure of TBP with Taf1 Reveals Anchoring Patterns in Transcriptional Regulation
Nat.Struct.Mol.Biol., 20, 2013
|
|
9FN2
 
 | | Crystal structure of the alkyltransferase ribozyme SAMURI co-crystallized with SAM | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, SAMURI-SAM | | Authors: | Chen, H.-A, Okuda, T, Lenz, A.-K, Scheitl, C.P.M, Schindelin, H, Hoebartner, C. | | Deposit date: | 2024-06-07 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and catalytic activity of the SAM-utilizing ribozyme SAMURI.
Nat.Chem.Biol., 2025
|
|
8GUJ
 
 | | Bre1-nucleosome complex (Model II) | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Onishi, S, Sato, K, Hamada, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
