2AN7
 
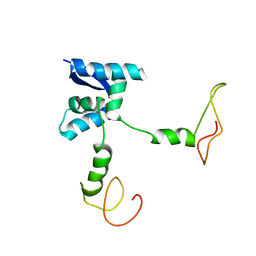 | | Solution structure of the bacterial antidote ParD | | 分子名称: | Protein parD | | 著者 | Oberer, M, Zangger, K, Gruber, K, Keller, W. | | 登録日 | 2005-08-11 | | 公開日 | 2006-09-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of ParD, the antidote of the ParDE toxin antitoxin module, provides the structural basis for DNA and toxin binding.
Protein Sci., 16, 2007
|
|
4WR8
 
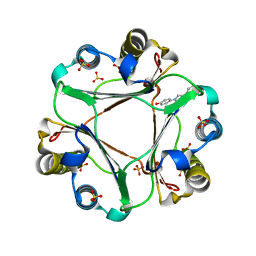 | | Macrophage Migration Inhibitory Factor in complex with a biaryltriazole inhibitor (3b-180) | | 分子名称: | 4-[4-(quinolin-2-yl)-1H-1,2,3-triazol-1-yl]phenol, Macrophage migration inhibitory factor, SODIUM ION, ... | | 著者 | Robertson, M.J, Baxter, R.H.G, Jorgensen, W.L. | | 登録日 | 2014-10-23 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Design, synthesis, and protein crystallography of biaryltriazoles as potent tautomerase inhibitors of macrophage migration inhibitory factor.
J.Am.Chem.Soc., 137, 2015
|
|
7UL5
 
 | |
7UL3
 
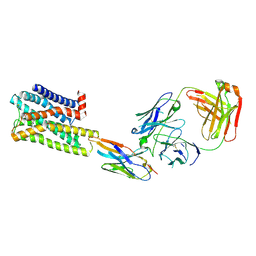 | | CryoEM Structure of Inactive H2R Bound to Famotidine, Nb6M, and NabFab | | 分子名称: | Histamine H2 receptor, NabFab HC, NabFab LC, ... | | 著者 | Robertson, M.J, Skiniotis, G. | | 登録日 | 2022-04-03 | | 公開日 | 2022-06-29 | | 最終更新日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UL2
 
 | |
7UL4
 
 | |
4WRB
 
 | | Macrophage Migration Inhibitory Factor in complex with a biaryltriazole inhibitor (3b-190) | | 分子名称: | 4-{4-[6-(2-methoxyethoxy)quinolin-2-yl]-1H-1,2,3-triazol-1-yl}phenol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Robertson, M.J, Baxter, R.H.G, Jorgensen, W.L. | | 登録日 | 2014-10-23 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Design, synthesis, and protein crystallography of biaryltriazoles as potent tautomerase inhibitors of macrophage migration inhibitory factor.
J.Am.Chem.Soc., 137, 2015
|
|
5J7Q
 
 | | Macrophage Migration Inhibitory Factor bound to Inhibitor K664 Derivative | | 分子名称: | 4-(imidazo[1,2-a]pyridin-2-yl)benzene-1,2-diol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Robertson, M.J, Jorgensen, W.L. | | 登録日 | 2016-04-06 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Irregularities in enzyme assays: The case of macrophage migration inhibitory factor.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5J7P
 
 | |
1P3H
 
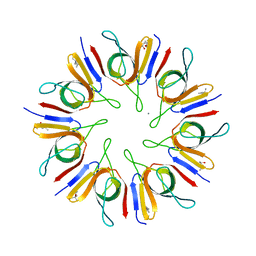 | | Crystal Structure of the Mycobacterium tuberculosis chaperonin 10 tetradecamer | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 10 kDa chaperonin, CALCIUM ION | | 著者 | Roberts, M.M, Coker, A.R, Fossati, G, Mascagni, P, Coates, A.R.M, Wood, S.P, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2003-04-17 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mycobacterium tuberculosis chaperonin 10 heptamers self-associate through their biologically active loops
J.BACTERIOL., 185, 2003
|
|
3ZNR
 
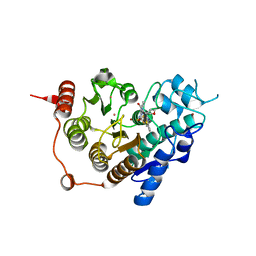 | | HDAC7 bound with inhibitor TMP269 | | 分子名称: | HISTONE DEACETYLASE 7, N-{[4-(4-phenyl-1,3-thiazol-2-yl)tetrahydro-2H-pyran-4-yl]methyl}-3-[5-(trifluoromethyl)-1,2,4-oxadiazol-3-yl]benzamide, POTASSIUM ION, ... | | 著者 | Lobera, m, madauss, k, pohlhaus, d, trump, r, nolan, m. | | 登録日 | 2013-02-15 | | 公開日 | 2013-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Selective Class Iia Histone Deacetylase Inhibition Via a Non-Chelating Zinc Binding Group
Nat.Chem.Biol., 9, 2013
|
|
3ZNS
 
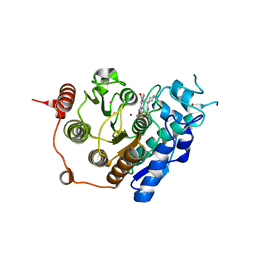 | | HDAC7 bound with TFMO inhibitor tmp942 | | 分子名称: | HISTONE DEACETYLASE 7, N-{[1-methyl-4-(4-phenyl-1,3-thiazol-2-yl)piperidin-4-yl]methyl}-3-[5-(trifluoromethyl)-1,2,4-oxadiazol-3-yl]benzamide, POTASSIUM ION, ... | | 著者 | Lobera, M, Madauss, K.P, Pohlhaus, D.T, Trump, R.P, Nolan, M.A. | | 登録日 | 2013-02-15 | | 公開日 | 2013-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Selective Class Iia Histone Deacetylase Inhibition Via a Non-Chelating Zinc Binding Group
Nat.Chem.Biol., 9, 2013
|
|
5HVT
 
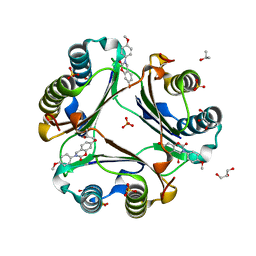 | | Crystal Structure of Macrophage Migration Inhibitory Factor (MIF) with a Potent Inhibitor (NVS-2) | | 分子名称: | 7-hydroxy-3-(4-methoxyphenyl)-3,4-dihydro-2H-1,3-benzoxazin-2-one, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Robertson, M.J, Jorgensen, W.L. | | 登録日 | 2016-01-28 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A Fluorescence Polarization Assay for Binding to Macrophage Migration Inhibitory Factor and Crystal Structures for Complexes of Two Potent Inhibitors.
J.Am.Chem.Soc., 138, 2016
|
|
6CBG
 
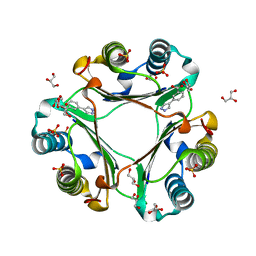 | | Macrophage Migration Inhibitory Factor in Complex with a Pyrazole Inhibitor (5) | | 分子名称: | 3-(1H-pyrazol-4-yl)benzoic acid, GLYCEROL, Macrophage migration inhibitory factor, ... | | 著者 | Robertson, M.J, Krimmer, S.G, Jorgensen, W.L. | | 登録日 | 2018-02-02 | | 公開日 | 2018-04-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Optimization of Pyrazoles as Phenol Surrogates to Yield Potent Inhibitors of Macrophage Migration Inhibitory Factor.
ChemMedChem, 13, 2018
|
|
6CBH
 
 | | Macrophage Migration Inhibitory Factor in Complex with a Pyrazole Inhibitor (8m) | | 分子名称: | 5-(3-fluoro-1H-pyrazol-4-yl)-2-[(naphthalen-2-yl)oxy]benzoic acid, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Robertson, M.J, Krimmer, S.G, Jorgensen, W.L. | | 登録日 | 2018-02-02 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Optimization of Pyrazoles as Phenol Surrogates to Yield Potent Inhibitors of Macrophage Migration Inhibitory Factor.
ChemMedChem, 13, 2018
|
|
5HVS
 
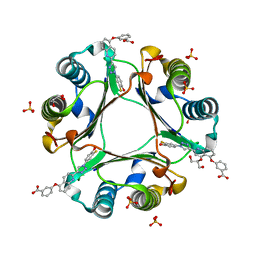 | | Crystal Structure of Macrophage Migration Inhibitory Factor (MIF) with a Biaryltriazole Inhibitor (3i-305) | | 分子名称: | 3-({2-[1-(3-fluoro-4-hydroxyphenyl)-1H-1,2,3-triazol-4-yl]quinolin-5-yl}oxy)benzoic acid, GLYCEROL, Macrophage migration inhibitory factor, ... | | 著者 | Robertson, M.J, Jorgensen, W.L. | | 登録日 | 2016-01-28 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A Fluorescence Polarization Assay for Binding to Macrophage Migration Inhibitory Factor and Crystal Structures for Complexes of Two Potent Inhibitors.
J.Am.Chem.Soc., 138, 2016
|
|
7T10
 
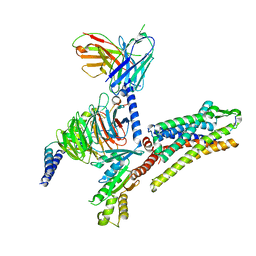 | |
7T11
 
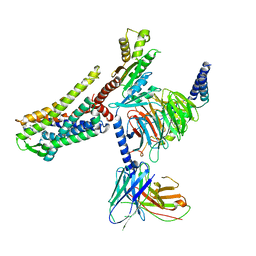 | |
6CBF
 
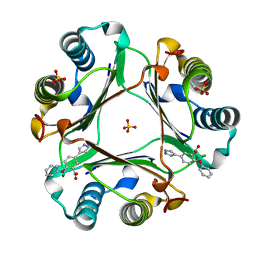 | |
1XJR
 
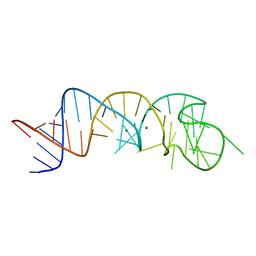 | | The Structure of a Rigorously Conserved RNA Element Within the SARS Virus Genome | | 分子名称: | MAGNESIUM ION, s2m RNA | | 著者 | Robertson, M.P, Igel, H, Baertsch, R, Haussler, D, Ares Jr, M, Scott, W.G. | | 登録日 | 2004-09-24 | | 公開日 | 2005-02-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The structure of a rigorously conserved RNA element within the SARS virus genome
Plos Biol., 3, 2005
|
|
2OIU
 
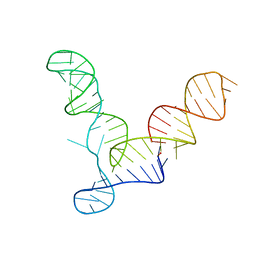 | |
1XC8
 
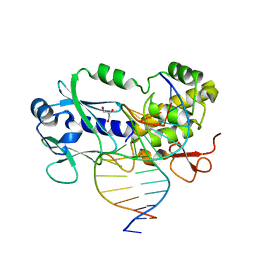 | | CRYSTAL STRUCTURE COMPLEX BETWEEN THE WILD-TYPE LACTOCOCCUS LACTIS FPG (MUTM) AND A FAPY-DG CONTAINING DNA | | 分子名称: | 5'-D(*CP*TP*CP*TP*TP*TP*(FOX)P*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', Formamidopyrimidine-DNA glycosylase, ... | | 著者 | Coste, F, Ober, M, Carell, T, Boiteux, S, Zelwer, C, Castaing, B. | | 登録日 | 2004-09-01 | | 公開日 | 2005-09-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase.
J.Biol.Chem., 279, 2004
|
|
6FL1
 
 | | Crystal structure of the complex between the Lactococcus lactis FPG mutant T221P and a Fapy-dG containing DNA | | 分子名称: | DNA (5'-D(*CP*TP*CP*TP*TP*TP(FOX)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | 著者 | Coste, F, Castaing, B, Ober, M, Carell, T. | | 登録日 | 2018-01-25 | | 公開日 | 2019-02-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of the complex between the Lactococcus lactis FPG mutant T221P and a Fapy-dG containing DNA
To Be Published
|
|
1U45
 
 | | 8oxoguanine at the pre-insertion site of the polymerase active site | | 分子名称: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | 著者 | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | 登録日 | 2004-07-23 | | 公開日 | 2004-09-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1U4B
 
 | | Extension of an adenine-8oxoguanine mismatch | | 分子名称: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | 著者 | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | 登録日 | 2004-07-23 | | 公開日 | 2004-09-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
