1JQB
 
 | | Alcohol Dehydrogenase from Clostridium Beijerinckii: Crystal Structure of Mutant with Enhanced Thermal Stability | | Descriptor: | NADP-dependent Alcohol Dehydrogenase, ZINC ION | | Authors: | Levin, I, Frolow, F, Bogin, O, Peretz, M, Hacham, Y, Burstein, Y. | | Deposit date: | 2001-08-05 | | Release date: | 2002-11-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the enhanced thermal stability of alcohol dehydrogenase
mutants from the mesophilic bacterium Clostridium beijerinckii:
contribution of salt bridging
Protein Sci., 11, 2002
|
|
1JT1
 
 | | FEZ-1 metallo-beta-lactamase from Legionella gormanii modelled with D-captopril | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, CHLORIDE ION, FEZ-1, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Papamicael, C, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2001-08-20 | | Release date: | 2003-01-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three-dimensional Structure of FEZ-1, a Monomeric Subclass B3 Metallo-[beta]-lactamase from Fluoribacter gormanii, in Native Form and in Complex with -Captopril
J.MOL.BIOL., 325, 2003
|
|
1OQF
 
 | | Crystal structure of the 2-methylisocitrate lyase | | Descriptor: | 2-methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Dunaway-Mariano, D, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-03-08 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 2-methylisocitrate lyase in complex with product and with isocitrate inhibitor provide insight into lyase substrate specificity, catalysis and evolution.
Biochemistry, 44, 2005
|
|
1JGI
 
 | |
1OTV
 
 | | PqqC, Pyrroloquinolinquinone Synthase C | | Descriptor: | Coenzyme PQQ synthesis protein C | | Authors: | Magnusson, O.T, Toyama, H, Saeki, M, Rojas, A, Reed, J.C, Adachi, O, Klinman, J.P, SChwarzenbacher, R. | | Deposit date: | 2003-03-23 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Quinone Biogenesis: Structure and Mechanism of PqqC, the Final Catalyst in the Production of Pyrroloquinoline Quinone.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6EMZ
 
 | |
1JOE
 
 | | Crystal Structure of Autoinducer-2 Production Protein (LuxS) from Heamophilus influenzae | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN, MERCURY (II) ION, ZINC ION | | Authors: | Chen, C.C.H, Parsons, J.F, Lim, K, Lehmann, C, Tempczyk, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-27 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF AUTOINDUCER-2 PRODUCTION PROTEIN (LUXS) FROM HEAMOPHILUS INFLUENZAE--A CASE OF TWINNED CRYSTAL
To be Published
|
|
1JF1
 
 | | Crystal structure of HLA-A2*0201 in complex with a decameric altered peptide ligand from the MART-1/Melan-A | | Descriptor: | HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-2 ALPHA CHAIN, ZINC ION, ... | | Authors: | Sliz, P, Michielin, O, Cerottini, J.C, Luescher, I, Romero, P, Karplus, M, Wiley, D.C. | | Deposit date: | 2001-06-19 | | Release date: | 2001-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of two closely related but antigenically distinct HLA-A2/melanocyte-melanoma tumor-antigen peptide complexes.
J.Immunol., 167, 2001
|
|
1KDR
 
 | | CYTIDINE MONOPHOSPHATE KINASE FROM E.COLI IN COMPLEX WITH ARA-CYTIDINE MONOPHOSPHATE | | Descriptor: | CYTIDYLATE KINASE, CYTOSINE ARABINOSE-5'-PHOSPHATE, SULFATE ION | | Authors: | Bertrand, T, Briozzo, P, Assairi, L, Ofiteru, A, Bucurenci, N, Munier-Lehmann, H, Golinelli-Pimpaneau, B, Barzu, O, Gilles, A.M. | | Deposit date: | 2001-11-13 | | Release date: | 2002-01-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sugar specificity of bacterial CMP kinases as revealed by crystal structures and mutagenesis of Escherichia coli enzyme.
J.Mol.Biol., 315, 2002
|
|
1P93
 
 | | CRYSTAL STRUCTURE OF THE AGONIST FORM OF GLUCOCORTICOID RECEPTOR | | Descriptor: | DEXAMETHASONE, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | Kauppi, B, Jakob, C, Farnegardh, M, Yang, J, Ahola, H, Alarcon, M, Calles, K, Engstrom, O, Harlan, J, Muchmore, S, Ramqvist, A.-K, Thorell, S, Ohman, L, Greer, J, Gustafsson, J.-A, Carlstedt-Duke, J, Carlquist, M. | | Deposit date: | 2003-05-09 | | Release date: | 2003-07-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Three-dimensional Structures of Antagonistic and Agonistic Forms of the Glucocorticoid Receptor Ligand-binding Domain:
RU-486 INDUCES A TRANSCONFORMATION THAT LEADS TO ACTIVE ANTAGONISM.
J.Biol.Chem., 278, 2003
|
|
1MJI
 
 | | DETAILED ANALYSIS OF RNA-PROTEIN INTERACTIONS WITHIN THE BACTERIAL RIBOSOMAL PROTEIN L5/5S RRNA COMPLEX | | Descriptor: | 50S ribosomal protein L5, 5S rRNA fragment, MAGNESIUM ION, ... | | Authors: | Perederina, A, Nevskaya, N, Nikonov, O, Nikulin, A, Dumas, P, Yao, M, Tanaka, I, Garber, M, Gongadze, G, Nikonov, S. | | Deposit date: | 2002-08-28 | | Release date: | 2003-01-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detailed analysis of RNA-protein interactions within the bacterial ribosomal
protein L5/5S rRNA complex
RNA, 8, 2002
|
|
1KO3
 
 | | VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa with Cys221 reduced | | Descriptor: | ACETATE ION, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Garcia-Saez, I, Docquier, J.-D, Rossolini, G.M, Dideberg, O. | | Deposit date: | 2001-12-20 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The three-dimensional structure of VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form
J.Mol.Biol., 375, 2008
|
|
1MPT
 
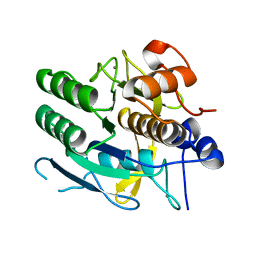 | | CRYSTAL STRUCTURE OF A NEW ALKALINE SERINE PROTEASE (M-PROTEASE) FROM BACILLUS SP. KSM-K16 | | Descriptor: | CALCIUM ION, M-PROTEASE | | Authors: | Yamane, T, Kani, T, Hatanaka, T, Suzuki, A, Ashida, T, Kobayashi, T, Ito, S, Yamashita, O. | | Deposit date: | 1994-04-13 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a new alkaline serine protease (M-protease) from Bacillus sp. KSM-K16.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
3H02
 
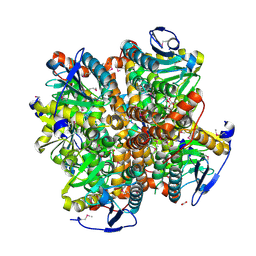 | | 2.15 Angstrom Resolution Crystal Structure of Naphthoate Synthase from Salmonella typhimurium. | | Descriptor: | BICARBONATE ION, Naphthoate synthase, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-08 | | Release date: | 2009-04-21 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Resolution Crystal Structure of Naphthoate Synthase from Salmonella typhimurium.
TO BE PUBLISHED
|
|
6F6R
 
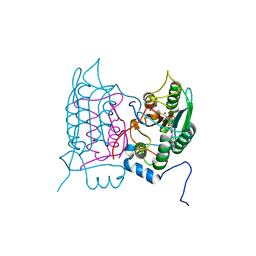 | | Crystal structure of human Caspase-1 with N-{3-[1-((S)-2-Hydroxy-5-oxo-tetrahydro-furan-3-ylcarbamoyl)-ethyl]-1-methyl-2,4-dioxo-1,2,3,4-tetrahydro-pyrimidin-5-yl}-4-(quinoxalin-2-ylamino)-benzamide | | Descriptor: | (3~{S})-3-[[(2~{R})-2-[3-methyl-2,6-bis(oxidanylidene)-5-[[4-(quinoxalin-2-ylamino)phenyl]carbonylamino]pyrimidin-1-yl]propanoyl]amino]-4-oxidanyl-butanoic acid, Caspase-1, SULFATE ION | | Authors: | Fournier, J.F, Clary, L, Chambon, S, Dumais, L, Harris, C.S, Millois-Barbuis, C, Pierre, R, Talano, S, Thoreau, E, Aubert, J, Aurelly, M, Bouix-Peter, C, Brethon, A, Chantalat, L, Christin, O, Comino, C, El-Bazbouz, G, Ghilini, A.L, Isabet, T, Lardy, C, Luzy, A.P, Mathieu, C, Mebrouk, K, Orfila, D, Pascau, J, Reverse, K, Roche, D, Rodeschini, V, Hennequin, L.F. | | Deposit date: | 2017-12-06 | | Release date: | 2018-05-02 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Drug Design of Topically Administered Caspase 1 Inhibitors for the Treatment of Inflammatory Acne.
J. Med. Chem., 61, 2018
|
|
6DUX
 
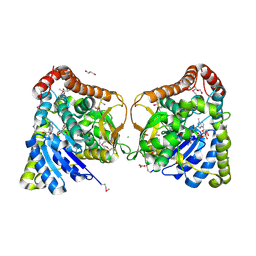 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD. | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 6-phospho-alpha-glucosidase, ACETATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-22 | | Release date: | 2018-07-04 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6DMS
 
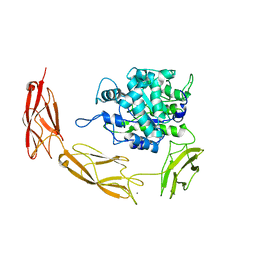 | | Endo-fucoidan hydrolase MfFcnA4_H294Q from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-05 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
1I6U
 
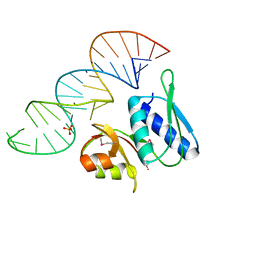 | | RNA-PROTEIN INTERACTIONS: THE CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN S8/RRNA COMPLEX FROM METHANOCOCCUS JANNASCHII | | Descriptor: | 16S RRNA FRAGMENT, 30S RIBOSOMAL PROTEIN S8P, SULFATE ION | | Authors: | Tishchenko, S, Nikulin, A, Fomenkova, N, Nevskaya, N, Nikonov, O, Dumas, P, Moine, H, Ehresmann, B, Ehresmann, C, Piendl, W, Lamzin, V, Garber, M, Nikonov, S. | | Deposit date: | 2001-03-05 | | Release date: | 2001-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Detailed analysis of RNA-protein interactions within the ribosomal protein S8-rRNA complex from the archaeon Methanococcus jannaschii.
J.Mol.Biol., 311, 2001
|
|
3H0C
 
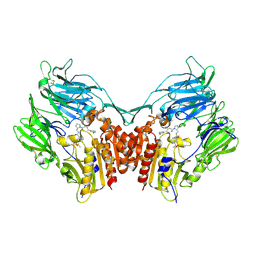 | | Crystal Structure of Human Dipeptidyl Peptidase IV (CD26) in Complex with a Reversed Amide Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, N-({(2S)-1-[(3R)-3-amino-4-(3-chlorophenyl)butanoyl]pyrrolidin-2-yl}methyl)-3-(methylsulfonyl)benzamide | | Authors: | Nordhoff, S, Cerezo-Galvez, S, Deppe, H, Hill, O, Lopez-Canet, M, Rummey, C, Thiemann, M, Matassa, V.G, Edwards, P.J, Feurer, A. | | Deposit date: | 2009-04-09 | | Release date: | 2009-06-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of b-homophenylalanine based pyrrolidin-2-ylmethyl amides and sulfonamides as highly potent and selective inhibitors of dipeptidyl peptidase IV
To be Published
|
|
6DWD
 
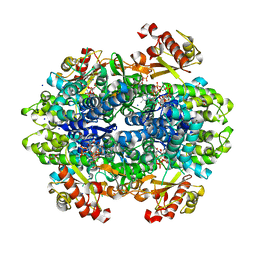 | | SAMHD1 Bound to Clofarabine-TP in the Catalytic Pocket and Allosteric Pocket | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, 9-{2-deoxy-2-fluoro-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}-2-me thyl-9H-purin-6-amine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1I5I
 
 | | THE C18S MUTANT OF BOVINE (GAMMA-B)-CRYSTALLIN | | Descriptor: | (GAMMA-B) CRYSTALLIN | | Authors: | Zarutskie, J.A, Asherie, N, Pande, J, Pande, A, Lomakin, J, Lomakin, A, Ogun, O, Stern, L.J, King, J.A, Benedek, G.B. | | Deposit date: | 2001-02-27 | | Release date: | 2001-03-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enhanced crystallization of the Cys18 to Ser mutant of bovine gammaB crystallin.
J.Mol.Biol., 314, 2001
|
|
3GP0
 
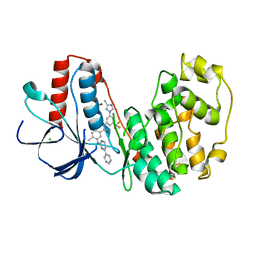 | | Crystal Structure of Human Mitogen Activated Protein Kinase 11 (p38 beta) in complex with Nilotinib | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Mitogen-activated protein kinase 11, ... | | Authors: | Filippakopoulos, P, Barr, A, Fedorov, O, Keates, T, Soundararajan, M, Elkins, J, Salah, E, Burgess-Brown, N, Ugochukwu, E, Pike, A.C.W, Muniz, J, Roos, A, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Mitogen Activated Protein Kinase 11 (p38 beta) in complex with Nilotinib
To be Published
|
|
6CWR
 
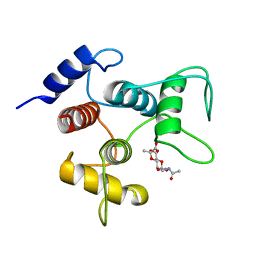 | | Crystal structure of SpaA-SLH/G46A/G109A in complex with 4,6-Pyr-beta-D-ManNAcOMe | | Descriptor: | Surface (S-) layer glycoprotein, methyl 2-(acetylamino)-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranoside | | Authors: | Blackler, R.J, Gagnon, S.M.L, Haji-Ghassemi, O, Evans, S.V. | | Deposit date: | 2018-03-30 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural basis of cell wall anchoring by SLH domains in Paenibacillus alvei.
Nat Commun, 9, 2018
|
|
3GNW
 
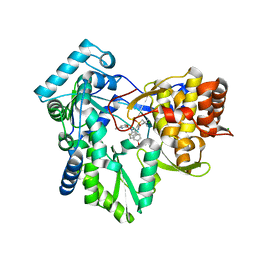 | | HCV NS5B polymerase in complex with 1,5 benzodiazepine inhibitor 4c | | Descriptor: | (11S)-11-[4-(benzyloxy)-2-fluorophenyl]-3,3-dimethyl-10-[(6-methylpyridin-2-yl)carbonyl]-2,3,4,5,10,11-hexahydrothiopyrano[3,2-b][1,5]benzodiazepin-6-ol 1,1-dioxide, CHLORIDE ION, RNA-directed RNA polymerase | | Authors: | Nyanguile, O, De Bondt, H. | | Deposit date: | 2009-03-18 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based design of a benzodiazepine scaffold yields a potent allosteric inhibitor of hepatitis C NS5B RNA polymerase.
J.Med.Chem., 52, 2009
|
|
3GUG
 
 | |
