7VR8
 
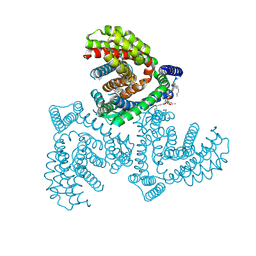 | | Inward-facing structure of human EAAT2 in the substrate-free state | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
7BTW
 
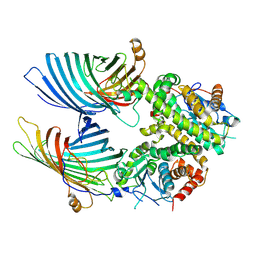 | | The mitochondrial SAM complex from S.cere | | Descriptor: | Mitochondrial outer membrane beta-barrel protein, SAM37 isoform 1, Sorting assembly machinery 35 kDa subunit | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7BTX
 
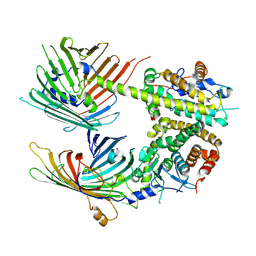 | | The mitochondrial SAM-Mdm10 supercomplex in GDN micelle from S.cere | | Descriptor: | MDM10 isoform 1, Mitochondrial outer membrane beta-barrel protein, Sorting assembly machinery 35 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7BTY
 
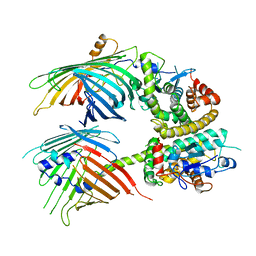 | | The mitochondrial SAM-Mdm10 supercomplex in Nanodisc from S.cere | | Descriptor: | MDM10 isoform 1, Mitochondrial outer membrane beta-barrel protein, Sorting assembly machinery 35 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7VTN
 
 | | Cryo-EM structure of the Cas13bt3-crRNA-target RNA ternary complex | | Descriptor: | Cas13bt3, crRNA, target RNA | | Authors: | Nakagawa, R, Soumya, K, Han, A, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Tomohiro, N, Yamashita, K, Feng, Z, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-10-30 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
8HIN
 
 | | Structure of human SGLT2-MAP17 complex with Phlorizin | | Descriptor: | 1-[2-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,6-bis(oxidanyl)phenyl]-3-(4-hydroxyphenyl)propan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, PDZK1-interacting protein 1, ... | | Authors: | Hiraizumi, M, Kishida, H, Miyaguchi, I, Nureki, O. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7V94
 
 | | Cryo-EM structure of the Cas12c2-sgRNA-target DNA ternary complex | | Descriptor: | Cas12c2, sgRNA, target DNA (non target strand), ... | | Authors: | Kurihara, N, Hirano, H, Tomita, A, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the type V-C CRISPR-Cas effector enzyme.
Mol.Cell, 82, 2022
|
|
7CCS
 
 | | Consensus mutated xCT-CD98hc complex | | Descriptor: | 4F2 cell-surface antigen heavy chain, Consensus mutated Anionic Amino Acid Transporter Light Chain, Xc- System | | Authors: | Oda, K, Lee, Y, Takemoto, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-06-17 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Consensus mutagenesis approach improves the thermal stability of system x c - transporter, xCT, and enables cryo-EM analyses.
Protein Sci., 29, 2020
|
|
4IKV
 
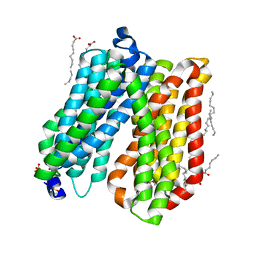 | | Crystal structure of peptide transporter POT | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Di-tripeptide ABC transporter (Permease), OLEIC ACID, ... | | Authors: | Doki, S, Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2012-12-28 | | Release date: | 2013-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7C7L
 
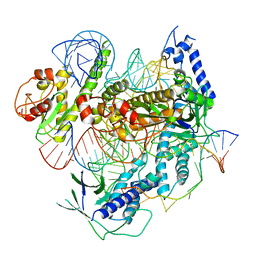 | | Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex | | Descriptor: | CRISPR-associated protein Cas14a.1, DNA (40-mer), ZINC ION, ... | | Authors: | Takeda, N.S, Nakagawa, R, Okazaki, S, Hirano, H, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the miniature type V-F CRISPR-Cas effector enzyme.
Mol.Cell, 81, 2021
|
|
4IKW
 
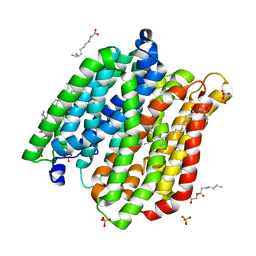 | | Crystal structure of peptide transporter POT in complex with sulfate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Di-tripeptide ABC transporter (Permease), OLEIC ACID, ... | | Authors: | Doki, S, Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2012-12-28 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IKY
 
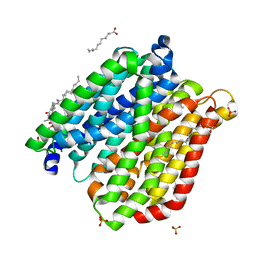 | | Crystal structure of peptide transporter POT (E310Q mutant) in complex with sulfate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Di-tripeptide ABC transporter (Permease), OLEIC ACID, ... | | Authors: | Doki, S, Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2012-12-28 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GTX
 
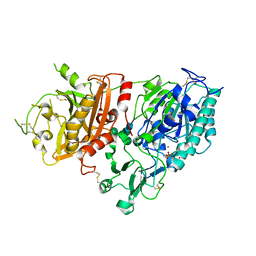 | | Crystal structure of mouse Enpp1 in complex with TMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5X2H
 
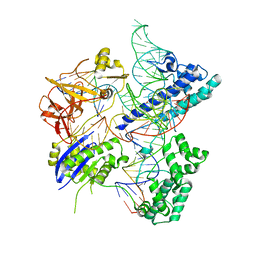 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5X2G
 
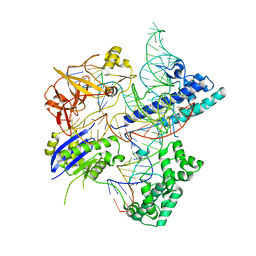 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
4GTW
 
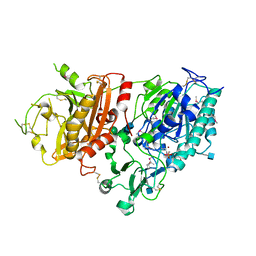 | | Crystal structure of mouse Enpp1 in complex with AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5ZSU
 
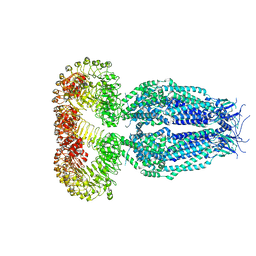 | | Structure of the human homo-hexameric LRRC8A channel at 4.25 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Kasuya, G, Nakane, T, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structures of the human volume-regulated anion channel LRRC8.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4GTZ
 
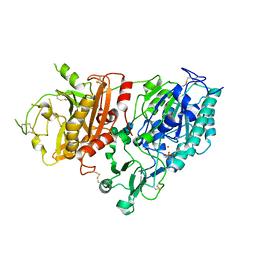 | | Crystal structure of mouse Enpp1 in complex with CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.191 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5X93
 
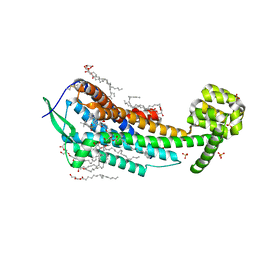 | | Human endothelin receptor type-B in complex with antagonist K-8794 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[6-[(4-tert-butylphenyl)sulfonylamino]-5-(2-methoxyphenoxy)-2-pyrimidin-2-yl-pyrimidin-4-yl]oxy-N-(2,6-dimethylphenyl)propanamide, CHOLESTEROL, ... | | Authors: | Shihoya, W, Nishizawa, T, Yamashita, K, Hirata, K, Okuta, A, Tani, K, Fujiyoshi, Y, Doi, T, Nureki, O. | | Deposit date: | 2017-03-05 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog
Nat. Struct. Mol. Biol., 24, 2017
|
|
4GTY
 
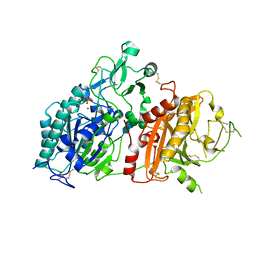 | | Crystal structure of mouse Enpp1 in complex with GMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5XH7
 
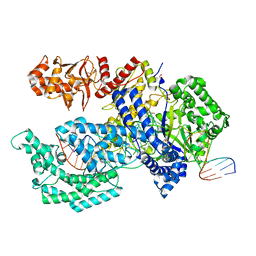 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RR variant in complex with crRNA and target DNA (TCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
4GEL
 
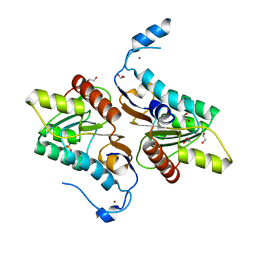 | | Crystal structure of Zucchini | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, PHOSPHATE ION, ... | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
2DB3
 
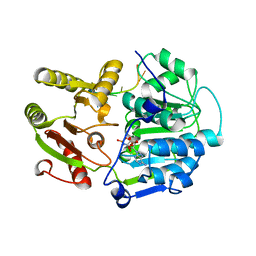 | | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase vasa, MAGNESIUM ION, ... | | Authors: | Sengoku, T, Nureki, O, Nakamura, A, Kobayashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.
Cell(Cambridge,Mass.), 125, 2006
|
|
4GEN
 
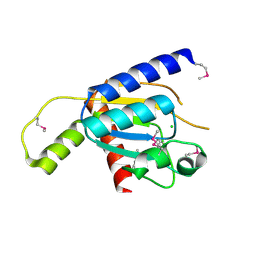 | | Crystal structure of Zucchini (monomer) | | Descriptor: | CHLORIDE ION, Mitochondrial cardiolipin hydrolase | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
7YO0
 
 | | Cryo-EM structure of human Slo1-LRRC26 complex with Symmetry Expansion | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CALCIUM ION, Calcium-activated potassium channel subunit alpha-1, ... | | Authors: | Yamanouchi, D, Nureki, O. | | Deposit date: | 2022-08-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for dual allosteric gating modulation of Slo1-LRRC channel complex
To Be Published
|
|
