6WV5
 
 | | Human VKOR C43S mutant with vitamin K1 epoxide | | Descriptor: | (2R,3R)-2-hydroxy-3-methyl-2-[(2E,7S)-3,7,11,15-tetramethylhexadec-2-en-1-yl]-2,3-dihydronaphthalene-1,4-dione, Vitamin K epoxide reductase Cys43Ser mutant, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
1R5Q
 
 | |
1R5W
 
 | | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T-cell activation | | Descriptor: | H-2 class II histocompatibility antigen, E-K alpha chain, MHC H2-IE-beta, ... | | Authors: | Krogsgaard, M, Prado, N, Adams, E, He, X, Chow, D.C, Wilson, D.B, Garcia, K.C, Davis, M.M. | | Deposit date: | 2003-10-13 | | Release date: | 2004-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation
Mol.Cell, 12, 2003
|
|
1R1K
 
 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to ponasterone A | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Billas, I.M.L, Iwema, T, Garnier, J.-M, Mitschler, A, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-24 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor.
Nature, 426, 2003
|
|
1BJ5
 
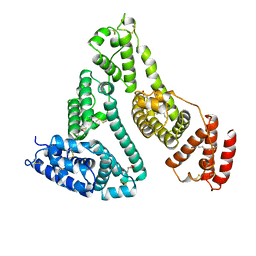 | | HUMAN SERUM ALBUMIN COMPLEXED WITH MYRISTIC ACID | | Descriptor: | HUMAN SERUM ALBUMIN, MYRISTIC ACID | | Authors: | Curry, S, Mandelkow, H, Brick, P, Franks, N. | | Deposit date: | 1998-07-02 | | Release date: | 1998-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites.
Nat.Struct.Biol., 5, 1998
|
|
8BF1
 
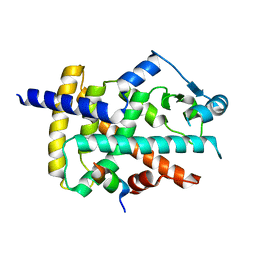 | |
5CUA
 
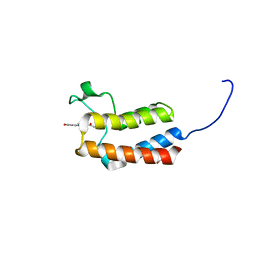 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 1-Acetyl-4-(4-hydroxyphenyl)piperazine (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(4-hydroxyphenyl)piperazin-1-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 1-Acetyl-4-(4-hydroxyphenyl)piperazine (SGC - Diamond I04-1 fragment screening)
To be published
|
|
6WHR
 
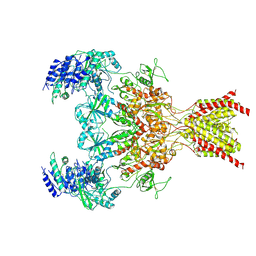 | | GluN1b-GluN2B NMDA receptor in non-active 2 conformation at 4 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
8BFF
 
 | | Human PPARgamma in complex with MINCH bound to the AF-2 sub-pocket | | Descriptor: | (1~{S},2~{R})-2-[(4~{R})-4-methylheptoxy]carbonylcyclohexane-1-carboxylic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Useini, A, Straeter, N. | | Deposit date: | 2022-10-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the activation of PPAR gamma by the plasticizer metabolites MEHP and MINCH.
Environ Int, 173, 2023
|
|
1BQF
 
 | | GROWTH-BLOCKING PEPTIDE (GBP) FROM PSEUDALETIA SEPARATA | | Descriptor: | PROTEIN (GROWTH-BLOCKING PEPTIDE) | | Authors: | Aizawa, T, Fujitani, N, Hayakawa, Y, Ohnishi, A, Ohkubo, T, Kwano, K, Hikichi, K, Nitta, K. | | Deposit date: | 1998-08-09 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an insect growth factor, growth-blocking peptide.
J.Biol.Chem., 274, 1999
|
|
1SM2
 
 | | Crystal structure of the phosphorylated Interleukin-2 tyrosine kinase catalytic domain | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C.M, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-08 | | Release date: | 2004-07-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
8BF2
 
 | |
1SIU
 
 | | KUMAMOLISIN-AS E78H MUTANT | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Tsuruoka, N, Ashida, M, Minakata, H, Oyama, H, Oda, K, Nishino, T, Nakayama, T. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-As, a serine-carboxyl peptidase with collagenase activity
J.Biol.Chem., 279, 2004
|
|
6CBC
 
 | |
5D4S
 
 | | Crystal Structure of AraR(DBD) in complex with operator ORX1 | | Descriptor: | Arabinose metabolism transcriptional repressor, DNA (5'-D(*AP*AP*AP*TP*AP*CP*AP*TP*AP*CP*GP*TP*AP*CP*AP*AP*AP*TP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*AP*TP*TP*TP*GP*TP*AP*CP*GP*TP*AP*TP*GP*TP*AP*TP*T)-3') | | Authors: | Jain, D, Narayanan, N, Nair, D.T. | | Deposit date: | 2015-08-08 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Plasticity in Repressor-DNA Interactions Neutralizes Loss of Symmetry in Bipartite Operators.
J.Biol.Chem., 291, 2016
|
|
1C1M
 
 | | PORCINE ELASTASE UNDER XENON PRESSURE (8 BAR) | | Descriptor: | CALCIUM ION, PROTEIN (PORCINE ELASTASE), SULFATE ION, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-07-22 | | Release date: | 1999-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
1SKU
 
 | | E. coli Aspartate Transcarbamylase 240's Loop Mutant (K244N) | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, MALONATE ION, ... | | Authors: | Alam, N, Stieglitz, K.A, Caban, M.D, Gourinath, S, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2004-03-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 240s Loop Interactions Stabilize the T State of Escherichia coli Aspartate Transcarbamoylase.
J.Biol.Chem., 279, 2004
|
|
5D4R
 
 | | Crystal Structure of AraR(DBD) in complex with operator ORE1 | | Descriptor: | Arabinose metabolism transcriptional repressor, DNA (5'-D(*AP*TP*AP*TP*TP*TP*GP*TP*AP*CP*GP*TP*AP*CP*TP*AP*AP*TP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*TP*AP*GP*TP*AP*CP*GP*TP*AP*CP*AP*AP*AP*TP*A)-3') | | Authors: | Jain, D, Narayanan, N, Nair, D.T. | | Deposit date: | 2015-08-08 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Plasticity in Repressor-DNA Interactions Neutralizes Loss of Symmetry in Bipartite Operators.
J.Biol.Chem., 291, 2016
|
|
4IRB
 
 | | Crystal Structure of Vaccinia Virus Uracil DNA Glycosylase Mutant del171-172D4 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Schormann, N, Zhukovskaya, N, Sartmatova, D, Nuth, M, Ricciardi, R.P, Chattopadhyay, D. | | Deposit date: | 2013-01-14 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations at the dimer interface affect both function and structure of the Vaccinia virus uracil DNA glycosylase
To be Published
|
|
4ITC
 
 | | Crystal Structure Analysis of the K1 Cleaved Adhesin domain of Lys-gingipain (Kgp) from Porphyromonas gingivalis W83 | | Descriptor: | CALCIUM ION, GLYCEROL, GUANIDINE, ... | | Authors: | Ganuelas, L.A, Li, N, Hunter, N, Collyer, C.A. | | Deposit date: | 2013-01-18 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The lysine gingipain adhesin domains from Porphyromonas gingivalis interact with erythrocytes and albumin: Structures correlate to function.
Eur J Microbiol Immunol (Bp), 3, 2013
|
|
1SR0
 
 | | Crystal structure of signalling protein from sheep(SPS-40) at 3.0A resolution using crystal grown in the presence of polysaccharides | | Descriptor: | beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, signal processing protein | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Singh, N, Kumar, J, Sharma, S, Singh, T.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of signalling protein from sheep(SPS-40) at 3.0A resolution using crystal grown in the presence of polysaccharides
To be Published
|
|
5EL7
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA and tRNALys in the A-site with a U-U mismatch in the second position and antibiotic paromomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2015-11-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
1SR9
 
 | | Crystal Structure of LeuA from Mycobacterium tuberculosis | | Descriptor: | 2-isopropylmalate synthase, 3-METHYL-2-OXOBUTANOIC ACID, CHLORIDE ION, ... | | Authors: | Koon, N, Squire, C.J, Baker, E.N. | | Deposit date: | 2004-03-22 | | Release date: | 2004-05-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of LeuA from Mycobacterium tuberculosis, a key enzyme in leucine biosynthesis
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6VAQ
 
 | |
7EHZ
 
 | | Structure of human NNMT in complex with macrocyclic peptide 2 | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide 2 | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
