7K5K
 
 | | Plasmodium vivax M17 leucyl aminopeptidase Pv-M17 | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, putative, ... | | Authors: | Malcolm, T.R, McGowan, S, Belousoff, M.J. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Active site metals mediate an oligomeric equilibrium in Plasmodium M17 aminopeptidases.
J.Biol.Chem., 296, 2020
|
|
6EE4
 
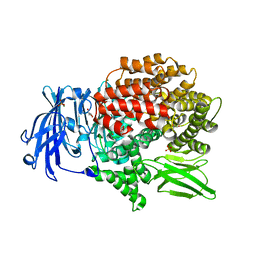 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6m) and catalytic zinc ion | | Descriptor: | (2R)-2-[(cyclopropylacetyl)amino]-N-hydroxy-2-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)acetamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
5J7C
 
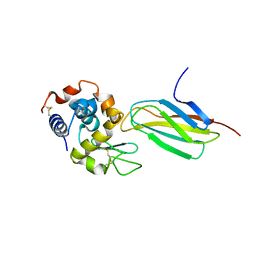 | |
5KYO
 
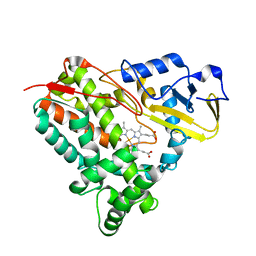 | | Crystal Structure of CYP101J2 | | Descriptor: | CYP101J2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Unterweger, B, Drinkwater, N, Dumsday, G.J, McGowan, S. | | Deposit date: | 2016-07-22 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of cytochrome P450 monooxygenase CYP101J2 from Sphingobium yanoikuyae strain B2.
Proteins, 85, 2017
|
|
6EE5
 
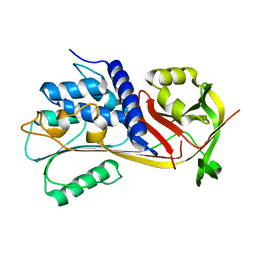 | |
6NVB
 
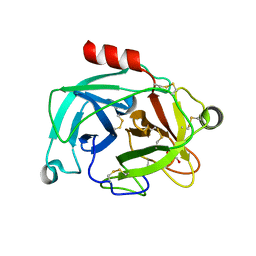 | | Crystal structure of the inhibitor-free form of the serine protease kallikrein-4 | | Descriptor: | GLYCEROL, Kallikrein-4, SULFATE ION | | Authors: | Riley, B.T, Buckle, A.M, McGowan, S. | | Deposit date: | 2019-02-04 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.636 Å) | | Cite: | Crystal structure of the inhibitor-free form of the serine protease kallikrein-4.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4Z0D
 
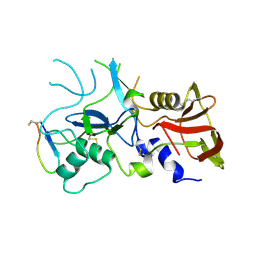 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Phe2038Trp] peptide | | Descriptor: | Apical membrane antigen 1, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4Z0E
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Phe2038TRN] peptide | | Descriptor: | Apical membrane antigen 1, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4Z0F
 
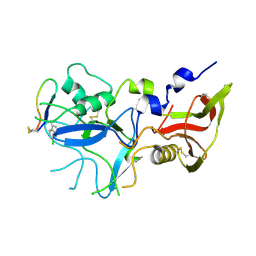 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Phe2038(6CW)] peptide | | Descriptor: | Apical membrane antigen 1, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4ZY1
 
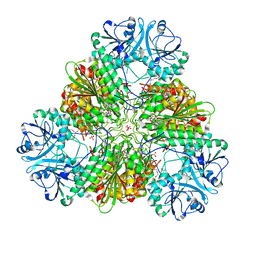 | |
4Z09
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Thr2040Ala] peptide | | Descriptor: | Apical membrane antigen 1, GLYCEROL, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4ZX3
 
 | |
5CDZ
 
 | | Crystal structure of conserpin in the latent state | | Descriptor: | Conserpin in the latent state, GLYCEROL | | Authors: | Porebski, B.T, McGowan, S, Keleher, S, Buckle, A.M. | | Deposit date: | 2015-07-06 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Smoothing a rugged protein folding landscape by sequence-based redesign.
Sci Rep, 6, 2016
|
|
4X2T
 
 | | X-ray crystal structure of the orally available aminopeptidase inhibitor, Tosedostat, bound to the M17 Leucyl Aminopeptidase from P. falciparum | | Descriptor: | (2S)-({(2R)-2-[(1S)-1-hydroxy-2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}amino)(phenyl)ethanoic acid, CARBONATE ION, M17 leucyl aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | X-ray crystal structures of an orally available aminopeptidase inhibitor, Tosedostat, bound to anti-malarial drug targets PfA-M1 and PfA-M17.
Proteins, 83, 2015
|
|
5CE0
 
 | |
5CBM
 
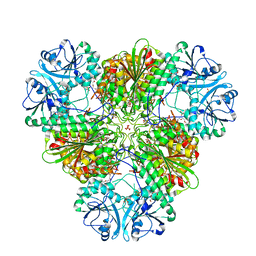 | | Crystal structure of PfA-M17 with virtual ligand inhibitor | | Descriptor: | (2S)-2-{[(R)-[(R)-amino(phenyl)methyl](hydroxy)phosphoryl]methyl}-4-methylpentanoic acid, CARBONATE ION, GLYCEROL, ... | | Authors: | Ruggeri, C, Drinkwater, N, McGowan, S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Validation of a Potent Dual Inhibitor of the P. falciparum M1 and M17 Aminopeptidases Using Virtual Screening.
Plos One, 10, 2015
|
|
5CDX
 
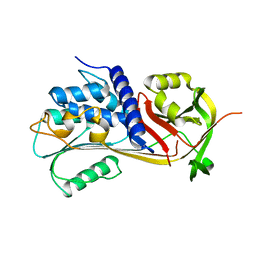 | |
4X2U
 
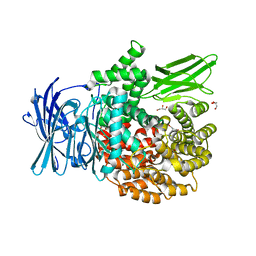 | | X-ray crystal structure of the orally available aminopeptidase inhibitor, Tosedostat, bound to the M1 Alanyl Aminopeptidase from P. falciparum | | Descriptor: | (2S)-({(2R)-2-[(1S)-1-hydroxy-2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}amino)(phenyl)ethanoic acid, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structures of an orally available aminopeptidase inhibitor, Tosedostat, bound to anti-malarial drug targets PfA-M1 and PfA-M17.
Proteins, 83, 2015
|
|
4WVM
 
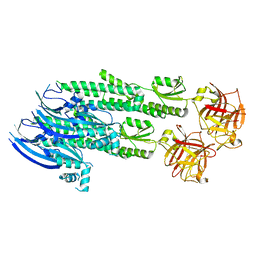 | | Stonustoxin structure | | Descriptor: | Stonustoxin subunit alpha, Stonustoxin subunit beta | | Authors: | Ellisdon, A.M, Panjikar, S, Whisstock, J.C, McGowan, S. | | Deposit date: | 2014-11-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Stonefish toxin defines an ancient branch of the perforin-like superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZQT
 
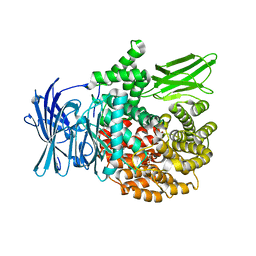 | | Crystal structure of PfA-M1 with virtual ligand inhibitor | | Descriptor: | (2R)-2-{[(R)-[(R)-amino(phenyl)methyl](hydroxy)phosphoryl]methyl}-4-methylpentanoic acid, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Ruggeri, C, Drinkwater, N, McGowan, S. | | Deposit date: | 2015-05-11 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Identification and Validation of a Potent Dual Inhibitor of the P. falciparum M1 and M17 Aminopeptidases Using Virtual Screening.
Plos One, 10, 2015
|
|
4R1B
 
 | |
4R1C
 
 | |
4R19
 
 | |
4R1A
 
 | |
4R5V
 
 | |
