7YV4
 
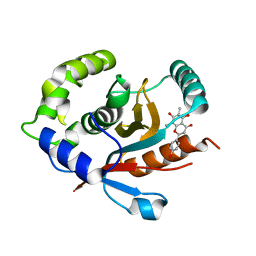 | | Crystal structure of human UCHL3 in complex with Farrerol | | Descriptor: | (2~{S})-2-(4-hydroxyphenyl)-6,8-dimethyl-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, Ubiquitin carboxyl-terminal hydrolase isozyme L3 | | Authors: | Mao, Z.Y, Xu, X.J, Zhang, W.T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Farrerol directly activates the deubiqutinase UCHL3 to promote DNA repair and reprogramming when mediated by somatic cell nuclear transfer.
Nat Commun, 14, 2023
|
|
9LWX
 
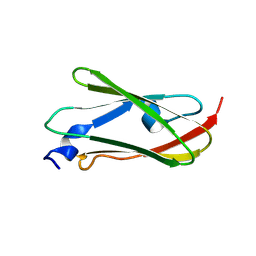 | |
9LXG
 
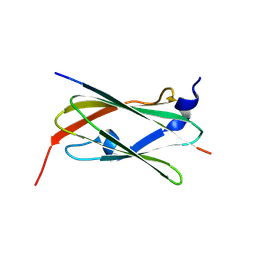 | |
7D64
 
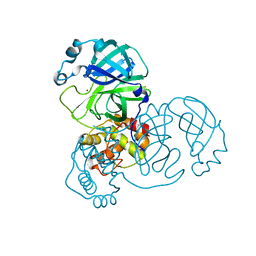 | |
9B6Z
 
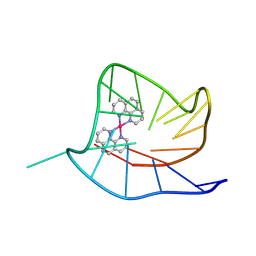 | |
8YU3
 
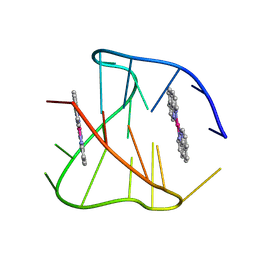 | |
7X2Z
 
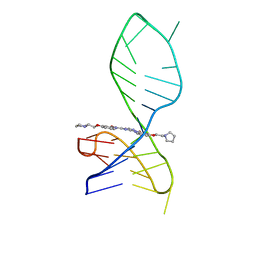 | | NMR solution structure of the 1:1 complex of a pyridostatin derivative (PyPDS) bound to a G-quadruplex MYT1L | | Descriptor: | 4-(2-azanylethoxy)-N2,N6-bis[4-(2-pyrrolidin-1-ylethoxy)quinolin-2-yl]pyridine-2,6-dicarboxamide, G-quadruplex DNA MYT1L | | Authors: | Liu, L.-Y, Mao, Z.-W, Liu, W. | | Deposit date: | 2022-02-26 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes.
J.Am.Chem.Soc., 144, 2022
|
|
6KFI
 
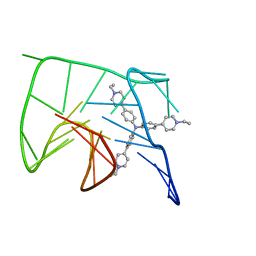 | | NMR solution structure of the 1:1 complex of Tel26 G-quadruplex and a tripodal cationic fluorescent probe NBTE | | Descriptor: | 4,4',4''-(nitrilotris(benzene-4,1-diyl))tris(1-ethylpyridin-1-ium) iodide, G-quadruplex DNA (26-MER) | | Authors: | Liu, W, Liu, L.Y, Mao, Z.W. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Quantitative Detection of G-Quadruplex DNA in Live Cells Based on Photon Counts and Complex Structure Discrimination.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6KFJ
 
 | | NMR solution structure of the 1:1 complex of wtTel26 G-quadruplex and a tripodal cationic fluorescent probe NBTE | | Descriptor: | 4,4',4''-(nitrilotris(benzene-4,1-diyl))tris(1-ethylpyridin-1-ium) iodide, G-quadruplex DNA wtTel26 | | Authors: | Liu, W, Liu, L.Y, Mao, Z.W. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Quantitative Detection of G-Quadruplex DNA in Live Cells Based on Photon Counts and Complex Structure Discrimination.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8IJC
 
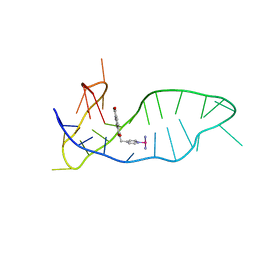 | | NMR solution structure of the 1:1 complex of a platinum(II) ligand L1-transpt covalently bound to a G-quadruplex MYT1L | | Descriptor: | G-quadruplex DNA MYT1L, Pt(NH3)2(2-(pyridin-4-ylmethyl)benzo-[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) | | Authors: | Liu, L.-Y, Mao, Z.-W. | | Deposit date: | 2023-02-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Organic-Platinum Hybrids for Covalent Binding of G-Quadruplexes: Structural Basis and Application to Cancer Immunotherapy.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7EL7
 
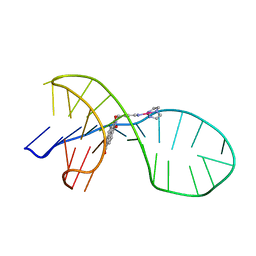 | | NMR solution structure of the 1:1 complex of a quadruplex-duplex hybrid MYT1L and a platinum(II) ligand L1Pt(dien) | | Descriptor: | G-quadruplex DNA MYT1L, Pt(diethylenetriamine)(2-(pyridin-4-ylmethyl)benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) | | Authors: | Liu, L.-Y, Liu, W, Mao, Z.-W. | | Deposit date: | 2021-04-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spatial Matching Selectivity and Solution Structure of Organic-Metal Hybrid to Quadruplex-Duplex Hybrid.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5Z80
 
 | | Solution structure for the 1:1 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | Descriptor: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | Authors: | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
5Z8F
 
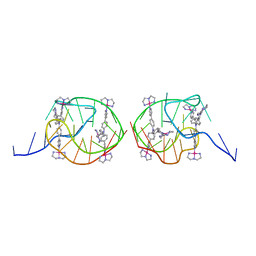 | | Solution structure for the unique dimeric 4:2 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | Descriptor: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | Authors: | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | Deposit date: | 2018-01-31 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
6LNZ
 
 | |
8GQD
 
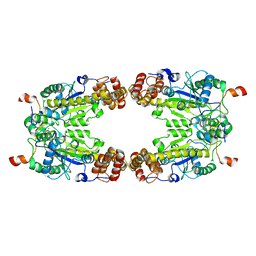 | | Complex Structure of Arginine Kinase McsB and McsA from Staphylococcus aureus | | Descriptor: | Protein-arginine kinase, Protein-arginine kinase activator protein, ZINC ION | | Authors: | Lu, K, Luo, B, Tao, X, Li, H, Xie, Y, Zhao, Z, Xia, W, Su, Z, Mao, Z. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Complex structure and activation mechanism of arginine kinase McsB by McsA.
Nat.Chem.Biol., 2024
|
|
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
8XKL
 
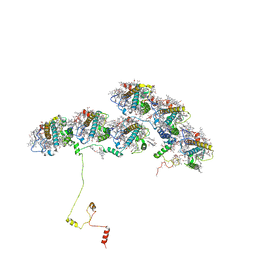 | | Structure of ACPII-CCPII from cryptophyte algae | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Li, X.Y, Mao, Z.Y, Shen, J.R, Han, G.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure and distinct supramolecular organization of a PSII-ACPII dimer from a cryptophyte alga Chroomonas placoidea.
Nat Commun, 15, 2024
|
|
7V3T
 
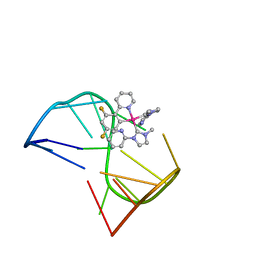 | | Solution structure of thrombin binding aptamer G-quadruplex bound a self-adaptive small molecule with rotated ligands | | Descriptor: | 11,13-bis(fluoranyl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-7$l^{4}-aza-8$l^{4}-platinatricyclo[7.4.0.0^{2,7}]trideca-1(9),2(7),3,5,10,12-hexaene, TBA G4 DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Liu, W, Zhu, B.C, Mao, Z.W. | | Deposit date: | 2021-08-11 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thrombin binding aptamer complex with a non-planar platinum(ii) compound.
Chem Sci, 13, 2022
|
|
8WB4
 
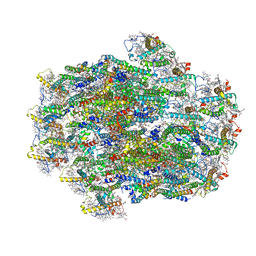 | | Structure of PSII-ACPII supercomplex from cryptophyte algae | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Li, X.Y, Mao, Z.Y, Shen, J.R, Han, G.Y. | | Deposit date: | 2023-09-08 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structure and distinct supramolecular organization of a PSII-ACPII dimer from a cryptophyte alga Chroomonas placoidea.
Nat Commun, 15, 2024
|
|
4NFT
 
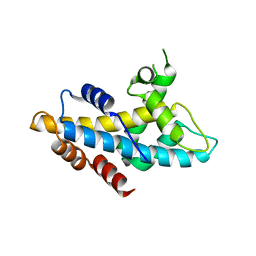 | | Crystal structure of human lnkH2B-h2A.Z-Anp32e | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member E, Histone H2B type 2-E, Histone H2A.Z | | Authors: | Shan, S, Pan, L, Mao, Z, Wang, W, Sun, J, Dong, Q, Liang, X, Ding, X, Chen, S, Dai, L, Zhang, Z, Zhu, B, Zhou, Z. | | Deposit date: | 2013-11-01 | | Release date: | 2014-04-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z
Cell Res., 24, 2014
|
|
7X3A
 
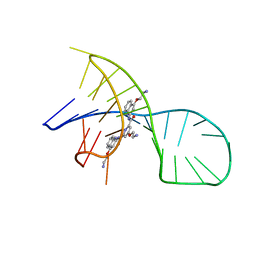 | | NMR solution structure of the 1:1 complex of a pyridostatin (PDS) bound to a G-quadruplex MYT1L | | Descriptor: | 4-(2-azanylethoxy)-N2,N6-bis[4-(2-azanylethoxy)quinolin-2-yl]pyridine-2,6-dicarboxamide, G-quadruplex DNA MYT1L | | Authors: | Liu, L.-Y, Mao, Z.-W, Liu, W. | | Deposit date: | 2022-02-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes.
J.Am.Chem.Soc., 144, 2022
|
|
