7ABG
 
 | | Human pre-Bact-1 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Cell division cycle 5-like protein, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7ABI
 
 | | Human pre-Bact-2 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Beta-catenin-like protein 1, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
4UYA
 
 | | Structure of MLK4 kinase domain with ATPgammaS | | Descriptor: | MAGNESIUM ION, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE MLK4, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Read, J.A, Brassington, C, Pollard, H.K, Phillips, C, Green, I, Overmann, R, Collier, M. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recurrent Mlk4 Loss-of-Function Mutations Suppress Jnk Signaling to Promote Colon Tumorigenesis.
Cancer Res., 76, 2016
|
|
3L9J
 
 | | Selection of a novel highly specific TNFalpha antagonist: Insight from the crystal structure of the antagonist-TNFalpha complex | | Descriptor: | MAGNESIUM ION, TNFalpha, Tumor necrosis factor, ... | | Authors: | Byla, P, Andersen, M.H, Thogersen, H.C, Gad, H.H, Hartmann, R. | | Deposit date: | 2010-01-05 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selection of a novel and highly specific TNF{alpha} antagonist: insight from the crystal structure of the antagonist-TNF{alpha} complex
J.Biol.Chem., 285, 2010
|
|
7AJT
 
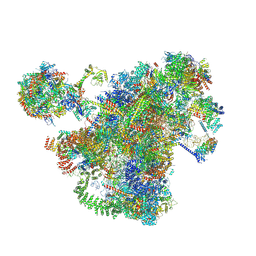 | | Cryo-EM structure of the 90S-exosome super-complex (state Pre-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
7B0Y
 
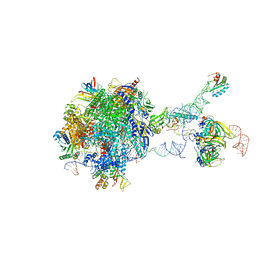 | | Structure of a transcribing RNA polymerase II-U1 snRNP complex | | Descriptor: | 145-nt RNA, DNA-directed RNA polymerase II subunit D, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Zhang, S, Aibara, S, Vos, S.M, Agafonov, D.E, Luehrmann, R, Cramer, P. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a transcribing RNA polymerase II-U1 snRNP complex.
Science, 371, 2021
|
|
1NM7
 
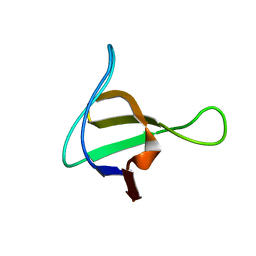 | | Solution structure of the ScPex13p SH3 domain | | Descriptor: | Peroxisomal Membrane Protein PAS20 | | Authors: | Pires, J.R, Hong, X, Brockmann, C, Volkmer-Engert, R, Schneider-Mergener, J, Oschkinat, H, Erdmann, R. | | Deposit date: | 2003-01-09 | | Release date: | 2003-03-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The ScPex13p SH3 Domain Exposes Two Distinct Binding Sites for Pex5p and Pex14p
J.Mol.Biol., 326, 2003
|
|
1JO6
 
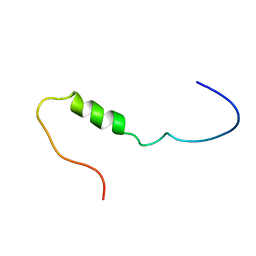 | | Solution structure of the cytoplasmic N-terminus of the BK beta-subunit KCNMB2 | | Descriptor: | potassium large conductance calcium-activated channel, subfamily M, beta member 2 | | Authors: | Bentrop, D, Beyermann, M, Wissmann, R, Fakler, B. | | Deposit date: | 2001-07-27 | | Release date: | 2001-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the "ball-and-chain" domain of KCNMB2, the beta 2-subunit of large conductance Ca2+- and voltage-activated potassium channels.
J.Biol.Chem., 276, 2001
|
|
3MSJ
 
 | | Structure of bace (beta secretase) in complex with inhibitor | | Descriptor: | 3-(2-amino-5-chloro-1H-benzimidazol-1-yl)propan-1-ol, BETA-SECRETASE 1, GLYCEROL | | Authors: | Madden, J, Kramer, J, Smith, M.A, Barker, J, Godemann, R. | | Deposit date: | 2010-04-29 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4XQ7
 
 | | The crystal structure of the OAS-like domain (OLD) of human OASL | | Descriptor: | 2'-5'-oligoadenylate synthase-like protein | | Authors: | Ibsen, M.S, Gad, H.H, Andersen, L.L, Hornung, V, Julkunen, I, Sarkar, S.N, Hartmann, R. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis reveals that human OASL binds dsRNA to enhance RIG-I signaling.
Nucleic Acids Res., 43, 2015
|
|
6ZON
 
 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 1 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZLW
 
 | | SARS-CoV-2 Nsp1 bound to the human 40S ribosomal subunit | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-01 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZN5
 
 | | SARS-CoV-2 Nsp1 bound to a pre-40S-like ribosome complex - state 2 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZP4
 
 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 2 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZVH
 
 | | EDF1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
6ZME
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-eERF1 ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZMT
 
 | | SARS-CoV-2 Nsp1 bound to a pre-40S-like ribosome complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZXH
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State H2 | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Ameismeier, M, Zemp, I, van den Heuvel, J, Thoms, M, Berninghausen, O, Kutay, U, Beckmann, R. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the final steps of human 40S ribosome maturation.
Nature, 587, 2020
|
|
6ZZZ
 
 | |
6ZQB
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state B2 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZXG
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State H1 | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Ameismeier, M, Zemp, I, van den Heuvel, J, Thoms, M, Berninghausen, O, Kutay, U, Beckmann, R. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for the final steps of human 40S ribosome maturation.
Nature, 587, 2020
|
|
7AAV
 
 | | Human pre-Bact-2 spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, D-chiro inositol hexakisphosphate, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7ZGO
 
 | | Cryo-EM structure of human NKCC1 (TM domain) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Nissen, P, Fenton, R, Neumann, C, Lindtoft Rosenbaek, L, Kock Flygaard, R, Habeck, M, Lykkegaard Karlsen, J, Wang, Y, Lindorff-Larsen, K, Gad, H, Hartmann, R, Lyons, J. | | Deposit date: | 2022-04-04 | | Release date: | 2022-10-05 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structure of the human NKCC1 transporter reveals mechanisms of ion coupling and specificity.
Embo J., 41, 2022
|
|
3MSK
 
 | | Fragment Based Discovery and Optimisation of BACE-1 Inhibitors | | Descriptor: | 4-(2-amino-5-chloro-1H-benzimidazol-1-yl)-N-cyclohexyl-N-methylbutanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Smith, M.A, Madden, J.M, Barker, J, Godemann, R, Kraemer, J, Hallett, D. | | Deposit date: | 2010-04-29 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1Q7X
 
 | | Solution structure of the alternatively spliced PDZ2 domain (PDZ2b) of PTP-Bas (hPTP1E) | | Descriptor: | PDZ2b domain of PTP-Bas (hPTP1E) | | Authors: | Kachel, N, Erdmann, K.S, Kremer, W, Wolff, P, Gronwald, W, Heumann, R, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-08-20 | | Release date: | 2003-12-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination and ligand interactions of the PDZ2b domain of PTP-Bas (hPTP1E): Splicing induced modulation of ligand specificity.
J.Mol.Biol., 334, 2003
|
|
