5AFW
 
 | | Assembly of methylated LSD1 and CHD1 drives AR-dependent transcription and translocation | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 1, ... | | 著者 | Metzger, E, Willmann, D, McMillan, J, Petroll, K, Metzger, P, Gerhardt, S, vonMaessenhausen, A, Schott, A.K, Espejo, A, Eberlin, A, Wohlwend, D, Schuele, K.M, Schleicher, M, Perner, S, Bedford, M.T, Dengjel, J, Flaig, R, Einsle, O, Schuele, R. | | 登録日 | 2015-01-26 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Assembly of Methylated Kdm1A and Chd1 Drives Androgen Receptor-Dependent Transcription and Translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3MJ4
 
 | | Crystal structure of UDP-galactopyranose mutase in complex with phosphonate analog of UDP-galactopyranose | | 分子名称: | (((2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)methyl)phosphonic (((2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl phosphoric) anhydride, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Karunan Partha, S, Sadeghi-Khomami, A, Slowski, K, Kotake, T, Thomas, N.R, Jakeman, D.L, Sanders, D.A.R. | | 登録日 | 2010-04-12 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Chemoenzymatic Synthesis, Inhibition Studies, and X-ray Crystallographic Analysis of the Phosphono Analog of UDP-Galp as an Inhibitor and Mechanistic Probe for UDP-Galactopyranose Mutase.
J.Mol.Biol., 403, 2010
|
|
6P0Z
 
 | | Crystal structure of N-acetylated KRAS (2-169) bound to GDP and Mg | | 分子名称: | ACETYL GROUP, DI(HYDROXYETHYL)ETHER, GTPase KRas, ... | | 著者 | Dharmaiah, S, Tran, T.H, Yan, W, Simanshu, D.K. | | 登録日 | 2019-05-17 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.011 Å) | | 主引用文献 | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
2CA0
 
 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | 分子名称: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Yermalitskaya, L.I, Kim, Y, Sherman, D.H, Waterman, M.R, Podust, L.M. | | 登録日 | 2005-12-15 | | 公開日 | 2006-12-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal Structure of Yc-17-Bound Cytochrome P450 Pikc (Cyp107L1)
To be Published
|
|
2CD8
 
 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | 分子名称: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Yermalitskaya, L.I, Kim, Y, Sherman, D.H, Waterman, M.R, Podust, L.M. | | 登録日 | 2006-01-20 | | 公開日 | 2007-02-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
3O7X
 
 | |
8FU3
 
 | | Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor | | 分子名称: | 8-methoxy-3-methyl-N-{(2S)-3,3,3-trifluoro-2-[5-fluoro-6-(4-fluorophenyl)-4-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-hydroxypropyl}cinnoline-6-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L | | 著者 | Yu, X, Abeywickrema, P, Bonneux, B, Behera, I, Jacoby, E, Fung, A, Adhikary, S, Bhaumik, A, Carbajo, R.J, Bruyn, S.D, Miller, R, Patrick, A, Pham, Q, Piassek, M, Verheyen, N, Shareef, A, Sutto-Ortiz, P, Ysebaert, N, Vlijmen, H.V, Jonckers, T.H.M, Herschke, F, McLellan, J.S, Decroly, E, Fearns, R, Grosse, S, Roymans, D, Sharma, S, Rigaux, P, Jin, Z. | | 登録日 | 2023-01-16 | | 公開日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor.
Commun Biol, 6, 2023
|
|
4Z18
 
 | | CRYSTAL STRUCTURE OF HUMAN PD-L1 | | 分子名称: | CHLORIDE ION, Programmed cell death 1 ligand 1 | | 著者 | Fedorov, A.A, Fedorov, E.V, Samantha, D, Hillerich, B, Seidel, R.D, Almo, S.C. | | 登録日 | 2015-03-27 | | 公開日 | 2015-04-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.952 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF HUMAN PD-L1
To Be Published
|
|
6GRB
 
 | | eukaryotic junction-resolving enzyme GEN-1 binding with Potassium | | 分子名称: | DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MAGNESIUM ION, ... | | 著者 | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | 登録日 | 2018-06-11 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
5AF6
 
 | | Structure of Lys33-linked diUb bound to Trabid NZF1 | | 分子名称: | TRABID, UBIQUITIN, ZINC ION | | 著者 | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | 登録日 | 2015-01-19 | | 公開日 | 2015-03-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
6PQ3
 
 | | Crystal structure of GDP-bound KRAS with ten residues long internal tandem duplication in the switch II region | | 分子名称: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Dharmaiah, S, Chan, A.H, Tran, T.H, Simanshu, D.K. | | 登録日 | 2019-07-08 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | RASinternal tandem duplication disrupts GTPase-activating protein (GAP) binding to activate oncogenic signaling.
J.Biol.Chem., 295, 2020
|
|
5A3F
 
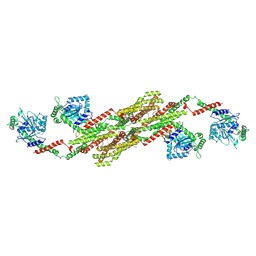 | | Crystal structure of the dynamin tetramer | | 分子名称: | DYNAMIN 3 | | 著者 | Reubold, T.F, Faelber, K, Plattner, N, Posor, Y, Branz, K, Curth, U, Schlegel, J, Anand, R, Manstein, D.J, Noe, F, Haucke, V, Daumke, O, Eschenburg, S. | | 登録日 | 2015-05-29 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Crystal Structure of the Dynamin Tetramer
Nature, 525, 2015
|
|
5C9V
 
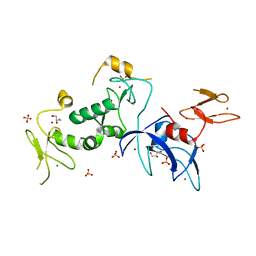 | | Structure of human Parkin G319A | | 分子名称: | E3 ubiquitin-protein ligase parkin, GLYCEROL, SULFATE ION, ... | | 著者 | Wauer, T, Komander, D. | | 登録日 | 2015-06-29 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Mechanism of phospho-ubiquitin-induced PARKIN activation.
Nature, 524, 2015
|
|
2HOR
 
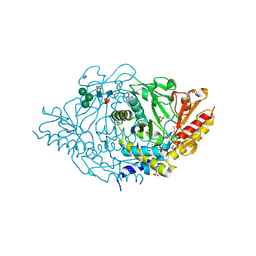 | | Crystal structure of alliinase from garlic- apo form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Alliin lyase 1, ... | | 著者 | Shimon, L.J.W, Rabinkov, A, Wilcheck, M, Mirelman, D, Frolow, F. | | 登録日 | 2006-07-16 | | 公開日 | 2007-02-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Two Structures of Alliinase from Alliium sativum L.: Apo Form and Ternary Complex with Aminoacrylate Reaction Intermediate Covalently Bound to the PLP Cofactor.
J.Mol.Biol., 366, 2007
|
|
3MXG
 
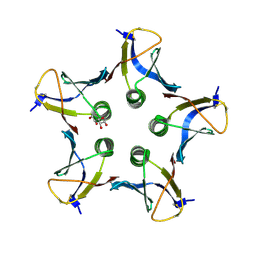 | | Structure of Shiga Toxin type 2 (Stx2) B Pentamer Mutant Q40L | | 分子名称: | D-xylose, Shiga-like toxin 2 subunit B | | 著者 | Kovall, R.A, Vander Wielen, B.D, Friedmann, D.R, Flagler, M.J. | | 登録日 | 2010-05-07 | | 公開日 | 2011-01-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Molecular basis of differential B-pentamer stability of Shiga toxins 1 and 2.
Plos One, 5, 2010
|
|
2KY5
 
 | | Solution structure of the PECAM-1 cytoplasmic tail with DPC | | 分子名称: | Platelet endothelial cell adhesion molecule | | 著者 | Lytle, B.L, Peterson, F.C, Volkman, B.F, Paddock, C, Newman, D.K, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2010-05-14 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Residues within a lipid-associated segment of the PECAM-1 cytoplasmic domain are susceptible to inducible, sequential phosphorylation.
Blood, 117, 2011
|
|
3LCR
 
 | | Thioesterase from Tautomycetin Biosynthhetic Pathway | | 分子名称: | DIMETHYL SULFOXIDE, FORMIC ACID, Tautomycetin biosynthetic PKS | | 著者 | Akey, D.L, Scaglione, J.B, Smith, J.L, Sherman, D.H. | | 登録日 | 2010-01-11 | | 公開日 | 2010-08-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Biochemical and structural characterization of the tautomycetin thioesterase: analysis of a stereoselective polyketide hydrolase.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2HOX
 
 | | alliinase from allium sativum (garlic) | | 分子名称: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Shimon, L.J.W, Rabinkov, A, Wilcheck, M, Mirelman, D, Frolow, F. | | 登録日 | 2006-07-17 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Two Structures of Alliinase from Alliium sativum L.: Apo Form and Ternary Complex with Aminoacrylate Reaction Intermediate Covalently Bound to the PLP Cofactor.
J.Mol.Biol., 366, 2007
|
|
3Q0D
 
 | | Crystal structure of SUVH5 SRA- hemi methylated CG DNA complex | | 分子名称: | CHLORIDE ION, DNA (5'-D(*CP*TP*GP*AP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*(5CM)P*GP*TP*CP*AP*G)-3'), ... | | 著者 | Eerappa, R, Simanshu, D.K, Patel, D.J. | | 登録日 | 2010-12-15 | | 公開日 | 2011-02-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3704 Å) | | 主引用文献 | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3TWZ
 
 | | Phosphorylated Bacillus cereus phosphopentomutase in space group P212121 | | 分子名称: | MANGANESE (II) ION, Phosphopentomutase | | 著者 | Panosian, T.P, Nanneman, D.P, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2011-09-22 | | 公開日 | 2012-02-29 | | 最終更新日 | 2012-03-21 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
3TX0
 
 | | Unphosphorylated Bacillus cereus phosphopentomutase in a P212121 crystal form | | 分子名称: | MANGANESE (II) ION, Phosphopentomutase | | 著者 | Panosian, T.P, Nanneman, D.P, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2011-09-22 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
7BLZ
 
 | | Red alga C.merolae Photosystem I | | 分子名称: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3R)-beta,beta-caroten-3-ol, ... | | 著者 | Nelson, N, Klaiman, D, Hippler, M. | | 登録日 | 2021-01-19 | | 公開日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Red alga C.merolae Photosystem I
To Be Published
|
|
7BNL
 
 | | Notum ARUK3003710 | | 分子名称: | (4~{E})-2-(3,4-dimethylphenyl)-4-[(1-methylpyrazol-4-yl)methylidene]-1,3-oxazol-5-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Y, Jones, E.Y, Fish, P.V, Svensson, F, Steadman, D. | | 登録日 | 2021-01-22 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | Notum Inhibitor ARUK3003710
To Be Published
|
|
6HEH
 
 | |
6HEI
 
 | |
