6G2H
 
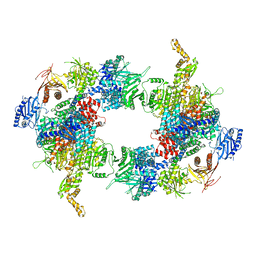 | | Filament of acetyl-CoA carboxylase and BRCT domains of BRCA1 (ACC-BRCT) core at 4.6 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
4ITF
 
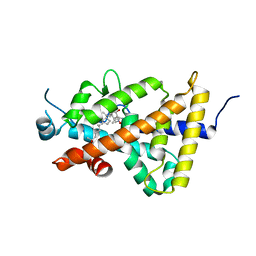 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 1alpha,25-Dihydroxy-2alpha-[2-(1H-tetrazole-1-yl)ethyl]vitamin D3 | | Descriptor: | (1R,2S,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methyl-6-oxidanyl-heptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-2-[2-(1,2,3,4-tetrazol-1-yl)ethyl]cyclohexane-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2013-01-18 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Synthesis of 2 alpha-heteroarylalkyl active vitamin d3 with therapeutic effect on enhancing bone mineral density in vivo
ACS MED.CHEM.LETT., 4, 2013
|
|
7PXW
 
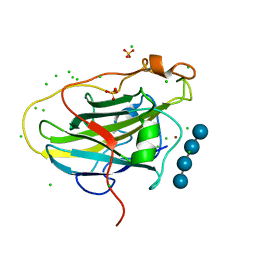 | | LPMO, expressed in E.coli, in complex with Cellotetraose | | Descriptor: | Auxiliary activity 9, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Banerjee, S, Muderspach, S.J, Tandrup, T, Ipsen, J, Rollan, C.H, Norholm, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
6G7H
 
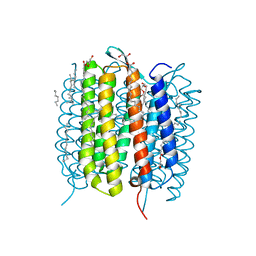 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: resting state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6G7Z
 
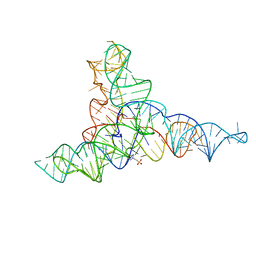 | |
8F6L
 
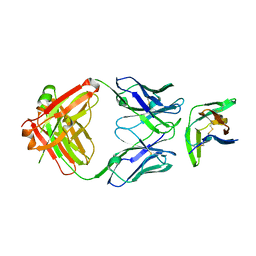 | | anti-BTLA monoclonal antibody h25F7 in complex with BTLA | | Descriptor: | B- and T-lymphocyte attenuator, h25F7 Fab heavy chain, h25F7 Fab light chain | | Authors: | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
7AQY
 
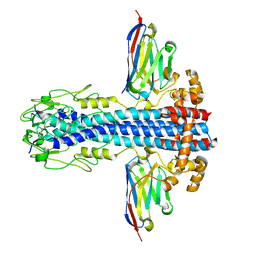 | |
8F2U
 
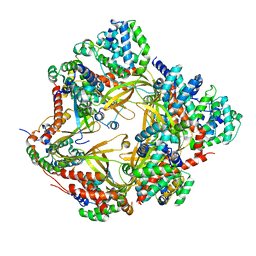 | | Human CCC complex | | Descriptor: | COMM domain-containing protein 1, COMM domain-containing protein 10, COMM domain-containing protein 2, ... | | Authors: | Healy, M.D, McNally, K.E, Butkovic, R, Chilton, M, Kato, K, Sacharz, J, McConville, C, Moody, E.R.R, Shaw, S, Planelles-Herrero, V.J, Kadapalakere, S.Y, Ross, J, Borucu, U, Palmer, C.S, Chen, K, Croll, T.I, Hall, R.J, Caruana, N.J, Ghai, R, Nguyen, T.H.D, Heesom, K.J, Saitoh, S, Berger, I, Berger-Schaffitzel, C, Williams, T.A, Stroud, D.A, Derivery, E, Collins, B.M, Cullen, P.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
3LB9
 
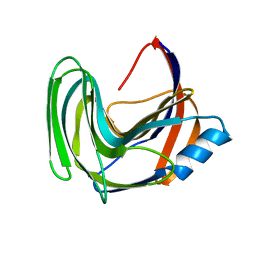 | | Crystal structure of the B. circulans cpA123 circular permutant | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | D'Angelo, I, Reitinger, S, Ludwiczek, M, Strynadka, N, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Circular permutation of Bacillus circulans xylanase: a kinetic and structural study.
Biochemistry, 49, 2010
|
|
5EMU
 
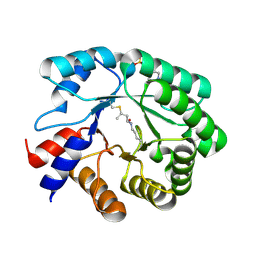 | |
7AQZ
 
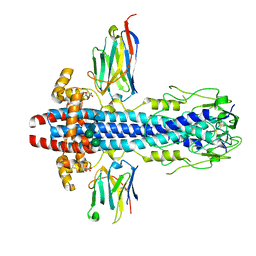 | | Co-Crystal Structure of Variant Surface Glycoprotein VSG2 in complex with Nanobody VSG2(NB14) | | Descriptor: | CITRIC ACID, Nanobody VSG2(NB14), SODIUM ION, ... | | Authors: | Stebbins, C.E, Hempelmann, A, VanStraaten, M. | | Deposit date: | 2020-10-23 | | Release date: | 2021-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Nanobody-mediated macromolecular crowding induces membrane fission and remodeling in the African trypanosome.
Cell Rep, 37, 2021
|
|
8ESM
 
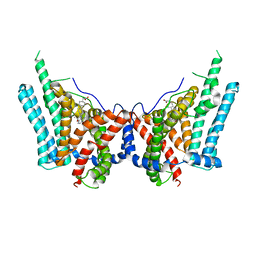 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with T863 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, {(1r,4r)-4-[4-(4-amino-7,7-dimethyl-7H-pyrimido[4,5-b][1,4]oxazin-6-yl)phenyl]cyclohexyl}acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-14 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
8ETM
 
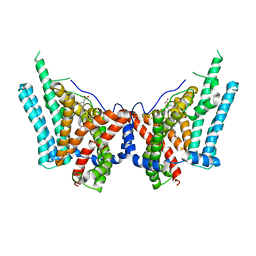 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with DGAT1IN1 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, [(1S,4r)-4-{4-[(4S)-2-({[4-(trifluoromethoxy)phenyl]methyl}carbamoyl)imidazo[1,2-a]pyridin-6-yl]phenyl}cyclohexyl]acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
3VT5
 
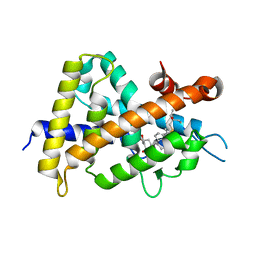 | | Crystal structures of rat VDR-LBD with R270L mutation | | Descriptor: | (1R,2E,3R,5Z,7E)-17-{(1S)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethylidene)-9,10-secoestra-5,7,16-triene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
7SK4
 
 | | Cryo-EM structure of ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ, an intracellular Fab, and an extracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK6
 
 | | Cryo-EM structure of human ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ and an intracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CID24 Fab heavy chain, CID24 Fab light chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK9
 
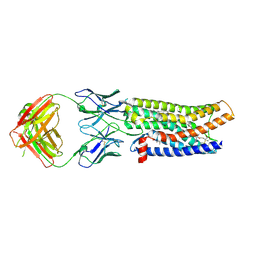 | | Cryo-EM structure of human ACKR3 in complex with a small molecule partial agonist CCX662, and an intracellular Fab | | Descriptor: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK3
 
 | | Cryo-EM structure of ACKR3 in complex with CXCL12, an intracellular Fab, and an extracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
3LK2
 
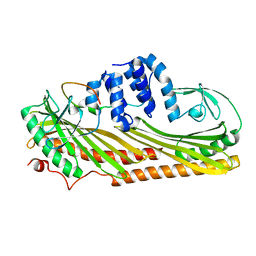 | | Crystal structure of CapZ bound to the uncapping motif from CARMIL | | Descriptor: | F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2, Leucine-rich repeat-containing protein 16A | | Authors: | Hernandez-Valladares, M, Kim, T, Kannan, B, Tung, A, Aguda, A.H, Larsson, M, Cooper, J.A, Robinson, R.C. | | Deposit date: | 2010-01-27 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6FOS
 
 | | Cyanidioschyzon merolae photosystem I | | Descriptor: | BETA-CAROTENE, CHLOROPHYLL A, IRON/SULFUR CLUSTER, ... | | Authors: | Nelson, N, Hippler, M, Antoshvili, M, Caspy, I. | | Deposit date: | 2018-02-08 | | Release date: | 2018-04-11 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure and function of photosystem I in Cyanidioschyzon merolae.
Photosyn. Res., 139, 2019
|
|
8P1V
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 6-cyclopropyl-~{N}-(2-methylindazol-5-yl)-1-propan-2-yl-pyrazolo[3,4-b]pyridine-4-carboxamide, CHLORIDE ION, ... | | Authors: | Thomsen, M, Thieulin-Pardo, G, Neumann, L. | | Deposit date: | 2023-05-12 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of novel methionine adenosyltransferase 2A (MAT2A) allosteric inhibitors by structure-based virtual screening.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
7SK8
 
 | | Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, an extracellular Fab, and an intracellular Fab | | Descriptor: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK5
 
 | | Cryo-EM structure of ACKR3 in complex with CXCL12 and an intracellular Fab | | Descriptor: | Anti-Fab nanobody, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
3LLH
 
 | | Crystal structure of the first dsRBD of TAR RNA-binding protein 2 | | Descriptor: | MALONATE ION, RISC-loading complex subunit TARBP2 | | Authors: | Yamashita, S, Kawazoe, M, Takemoto, C, Sekine, S, Wakiyama, M, Yokoyama, S. | | Deposit date: | 2010-01-29 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The structures of dsRBDs of human TRBP
To be Published
|
|
3LIZ
 
 | | crystal structure of bla g 2 complexed with Fab 4C3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C3 monoclonal antibody Heavy Chain, ... | | Authors: | Li, M, Gustchina, A, Glesner, J, Wunschmann, S, Pomes, A, Wlodawer, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of allergen-antibody interaction of cockroach allergen Bla g 2 with monoclonal antibodies that inhibit IgE antibody binding.
Plos One, 6, 2011
|
|
